class: center, middle, inverse, title-slide # Model Complications ## Sociology 513 ### Aaron Gullickson ### University of Oregon ### 2020-04-03 --- ## Jump to... [Linear Model, Revisited](#lm-revisited) [Nonlinearity](#nonlinearity) [IID Violations](#iid) [Sample Design and Weighting](#sample-design) [Missing Data](#missing-data) [Multicollinearity and Scale Creation](#multicollinearity) [Model Selection](#model-selection) --- class: inverse, center, middle background-image: url(images/jeremy-bishop-FzrlPh20l7Q-unsplash.jpg) background-size: cover name: lm-revisited # The Linear Model Revisited --- ## Review of linear model `$$\hat{y}_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\ldots+\beta_px_{ip}$$` -- - `\(\hat{y}_i\)` is the predicted value of the outcome/dependent variable which must be quantitative. -- - The `\(x\)` values are different independent/explanatory variables which may be quantitative or categorical (by using dummy coding). -- - The `\(\beta\)` values are the slopes/effects/coefficients that, *holding all other independent variables constant*, tell us the: - The predicted change in `\(y\)` for a one unit increase in `\(x\)` if `\(x\)` is quantitative - The mean difference in `\(y\)` between the indicated category and the reference category if `\(x\)` is categorical. -- ### Interpret these values ```r coef(lm(indegree~nsports+I(parentinc-45)+sex, data=popularity)) ``` ``` ## (Intercept) nsports I(parentinc - 45) sex Male ## 4.29086316 0.50668467 0.01131066 -0.62250550 ``` --- ## The residual term `$$\epsilon_i=\hat{y}_i-y_i$$` The residual/error term `\(\epsilon_i\)` gives us the difference between the actual value of the outcome variable for a given observation and the value predicted by the model. Lets use algebra to rewrite this equation with `\(y_i\)` on the left-hand side: -- `$$y_i=\hat{y}_i+\epsilon_i$$` If we plug in our linear model formula for `\(\hat{y}_i\)`, we can get the full model formula: -- `$$y_i=\underbrace{\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\ldots+\beta_px_{ip}}_{\hat{y}_i}+\epsilon_i$$` --- ## Linear model as a partition of the variance in `\(y\)` `$$y_i = \underbrace{\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\ldots+\beta_px_{ip}}_\text{structural} + \underbrace{\epsilon_i}_\text{stochastic}$$` - The structural part is the predicted value from our model which is typically a linear function of the independent variables. - The stochastic component is the leftover residual or error component, that is not accounted for by the model. -- Depending on disciplinary norms, there are different conceptual ways to view this basic relationship: -- - **Description:** observed = summary + residual -- - **Prediction:** observed = predicted + error -- - **Causation:** observed = true process + disturbance --- ## Calculating marginal effects The **marginal effect** of `\(x\)` on `\(y\)` is simply the predicted change in `\(y\)` for a one unit increase in `\(x\)` from its current value, holding all else constant. Sound familiar? -- In a basic linear model, the marginal effect is just given by the slope itself. `$$y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\epsilon_i$$` - `\(\beta_1\)` is the marginal effect of `\(x_1\)` - `\(\beta_2\)` is the marginal effect of `\(x_2\)` -- Technically, the marginal effect is the derivative of `\(y\)` with respect to a given `\(x\)`. This gives us the **tangent line** for the curve at any value of `\(x\)`. `$$\frac{\partial y}{\partial x_1}=\beta_1$$` `$$\frac{\partial y}{\partial x_2}=\beta_2$$` 😱 I am not expecting you to know the calculus behind this, but it may help some people --- ## Marginal effects with interaction terms .pull-left[ Marginal effects can get more complicated when we move to more complex model. Consider this model with an interaction term: `$$y_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\beta_3(x_{i1})(x_{i2})+\epsilon_i$$` -- The marginal effects are now given by: `$$\frac{\partial y}{\partial x_1}=\beta_1+\beta_3x_{i2}$$` `$$\frac{\partial y}{\partial x_2}=\beta_2+\beta_3x_{i1}$$` This is a very math-y way of saying: **the main effects depend on the effect of the other variable**. ] -- .pull-right[ ```r model <- lm(indegree~nsports*sex, data=popularity) as.data.frame(coef(model)) ``` ``` ## coef(model) ## (Intercept) 4.21374011 ## nsports 0.59169144 ## sex Male -0.54255556 ## nsports:sex Male -0.08760095 ``` `$$\hat{y}_i=4.21+0.49(nsports_i)-0.54(male_i)-0.09(nsports_i)(male_i)$$` The marginal effect for number of sports played is: `$$0.49-0.09(male_i)$$` The marginal effect for gender is: `$$-0.54-0.09(nsports_i)$$` ] --- ## Marginal effects plot .pull-left[ ```r nsports <- 0:6 gender_diff <- -0.54-0.09*nsports ggplot(data.frame(nsports, gender_diff), aes(x=nsports, y=gender_diff))+ geom_line()+ labs(x="number of sports played", y="predicted popularity difference between boys and girls") ``` ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-9-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Two key assumptions of linear models -- .pull-left[ ### Linearity <img src="module6_slides_model_complications_files/figure-html/linear-violation-1.png" width="504" style="display: block; margin: auto;" /> * If you fit a model with the wrong functional form, it is considered a *specification error*. * We can correct this through a variety of more advanced model specifications. ] -- .pull-right[ ### Error terms are iid <img src="module6_slides_model_complications_files/figure-html/heteroskedasticity-1.png" width="504" style="display: block; margin: auto;" /> * iid = **independent and identically distributed** which is typically violated either by **heteroscedasticity** or **autocorrelation**. * The consequence of violating the i.i.d. assumption is usually incorrect standard errors. ] --- ## How are linear model parameters estimated? ⚠️ Heavy math ahead! - We have simple formulas for the slope and intercept for the case of a single independent variable. - With multiple independent variables, a simple formula will not suffice. To estimate model parameters with multiple independent variables we need to use some matrix algebra. -- ### The matrix algebra approach to linear models We can use matrix algebra to represent our linear regression model equation using one-dimensional **vectors** and two-dimensional **matrices**. `$$\mathbf{y}=\mathbf{X\beta+\epsilon}$$` - `\(\mathbf{y}\)` is a vector of known values of the independent variable of length `\(n\)`. - `\(\mathbf{X}\)` is a matrix of known values of the independent variables of dimensions `\(n\)` by `\(p+1\)`. - `\(\mathbf{\beta}\)` is a vector of to-be-estimated values of intercepts and slopes of length `\(p+1\)`. - `\(\mathbf{\epsilon}\)` is a vector of residuals of length `\(n\)` that will be equal to `\(\mathbf{y-X\beta}\)`. --- ## ⚠️ Linear model in matrix form .pull-left[ `$$\mathbf{y}=\begin{pmatrix} y_{1}\\ y_{2}\\ \vdots\\ y_{n}\\ \end{pmatrix}$$` `$$\mathbf{X}=\begin{pmatrix} 1 & x_{11} & x_{12} & \ldots & x_{1p}\\ 1 & x_{21} & x_{22} & \ldots & x_{2p}\\ \vdots & \vdots & \ldots & \vdots\\ 1 & x_{n1} & x_{n2} & \ldots & x_{np}\\ \end{pmatrix}$$` ] .pull-right[ `$$\mathbf{\beta}=\begin{pmatrix} \beta_{1}\\ \beta_{2}\\ \vdots\\ \beta_{p}\\ \end{pmatrix}$$` `$$\mathbf{\epsilon}=\begin{pmatrix} \epsilon_{1}\\ \epsilon_{2}\\ \vdots\\ \epsilon_{n}\\ \end{pmatrix}$$` ] -- Putting it all together gives us this beast 👹: `$$\begin{pmatrix} y_{1}\\ y_{2}\\ \vdots\\ y_{n}\\ \end{pmatrix}=\begin{pmatrix} 1 & x_{11} & x_{12} & \ldots & x_{1p}\\ 1 & x_{21} & x_{22} & \ldots & x_{2p}\\ \vdots & \vdots & \ldots & \vdots\\ 1 & x_{n1} & x_{n2} & \ldots & x_{np}\\ \end{pmatrix}\begin{pmatrix} \beta_{1}\\ \beta_{2}\\ \vdots\\ \beta_{p}\\ \end{pmatrix}+\begin{pmatrix} \epsilon_{1}\\ \epsilon_{2}\\ \vdots\\ \epsilon_{n}\\ \end{pmatrix}$$` --- ## ⚠️ Minimize sum of squred residuals Our goal is to choose a `\(\mathbf{\beta}\)` vector that minimizes the sum of squared residuals, `\(SSR\)`, which is just given by the `\(\epsilon\)` vector squared and summed up. We can rewrite the matrix algebra formula to isolate `\(e\)` on one side: `$$\begin{align*} \mathbf{y}&=&\mathbf{X\beta+\epsilon}\\ \epsilon&=&\mathbf{y}-\mathbf{X\beta} \end{align*}$$` -- In matrix form, `\(SSR\)` can be represented as a function of `\(\mathbf{\beta}\)`, like so: `$$\begin{align*} SSR(\beta)&=&(\mathbf{y}-\mathbf{X\beta})'(\mathbf{y}-\mathbf{X\beta})\\ &=&\mathbf{y}'\mathbf{y}-2\mathbf{y}'\mathbf{X\beta}+\mathbf{\beta}'\mathbf{X'X\beta} \end{align*}$$` -- We want to choose a `\(\mathbf{\beta}\)` to plug into this function that provides the smallest possible value (the minimum). It turns out that we can get this value by using calculus to get the derivative with respect to `\(\mathbf{\beta}\)` and solving for zero: `$$0=-2\mathbf{X'y}+2\mathbf{X'X\beta}$$` -- Applying some matrix algebra will give us the estimator of `\(\mathbf{\beta}\)`: `$$\mathbf{\beta}=(\mathbf{X'X})^{-1}\mathbf{X'y}$$` --- ## Estimating linear models manually <div class="footer"> <body>Sociology 513, Model Complications: The Linear Model, Revisited</body> </div> ```r #set up the design matrix X <- as.matrix(cbind(rep(1, nrow(movies)), movies[,c("Runtime","BoxOffice")])) #the outcome variable vector y <- movies$TomatoMeter #crossprod(X) will do matrix multiplication, solve will invert beta <- solve(crossprod(X))%*%crossprod(X,y) beta ``` ``` ## [,1] ## rep(1, nrow(movies)) 11.09207358 ## Runtime 0.32320152 ## BoxOffice 0.05923506 ``` ```r #how does it compare to lm? model <- lm(TomatoMeter~Runtime+BoxOffice, data=movies) coef(model) ``` ``` ## (Intercept) Runtime BoxOffice ## 11.09207358 0.32320152 0.05923506 ``` It works! --- ## ⚠️ Estimating standard errors using matrix algebra If we treat `\(\sigma^2\)` as the variance of the error term `\(\epsilon\)`, then we can also use matrix algebra to calculate the **covariance matrix**: `$$\sigma^{2}(\mathbf{X'X})^{-1}$$` The values of this matrix give us information about the correlation between different independent variables. Most importantly, the square root of the diagonal values of this matrix are the standard errors for the estimated values of `\(\beta\)`. -- In practice, we don't have `\(\sigma^2\)`, but we can estimate from the fitted values of `\(y\)` by: `$$s^2=\frac{\sum(y_i-\hat{y}_i)^2}{n-p-1}$$` We can then use these estimated standard errors to calculate t-statistics and p-values, confidence intervals, and so on. --- ## Calculating SEs manually ```r y.hat <- X%*%beta df <- length(y)-ncol(X) s.sq <- sum((y-y.hat)^2)/df covar.matrix <- s.sq*solve(crossprod(X)) se <- sqrt(diag(covar.matrix)) t.stat <- beta/se p.value <- 2*pt(-1*abs(t.stat), df) data.frame(beta,se,t.stat,p.value) ``` ``` ## beta se t.stat p.value ## rep(1, nrow(movies)) 11.09207358 3.269128357 3.392976 7.019316e-04 ## Runtime 0.32320152 0.031734307 10.184609 6.617535e-24 ## BoxOffice 0.05923506 0.007978845 7.424014 1.540215e-13 ``` ```r summary(model)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 11.09207358 3.269128357 3.392976 7.019316e-04 ## Runtime 0.32320152 0.031734307 10.184609 6.617535e-24 ## BoxOffice 0.05923506 0.007978845 7.424014 1.540215e-13 ``` --- class: inverse, center, middle background-image: url(images/fabian-quintero-nq02weVF_mk-unsplash.jpg) background-size: cover name: nonlinearity # Modeling Non-Linearity --- ## Linear models fit straight lines <img src="module6_slides_model_complications_files/figure-html/life-exp-non-linear-1.png" width="864" style="display: block; margin: auto;" /> ??? - If you try to model a relationship using a straight line when that relationship is more complex (i.e. "bendy") then your model will not fit well and will not represent the relationship accurately. - The technical way of saying this is that the functional form of the relationship is mis-specified in the model. - The most common cases of non-linearity are **diminishing returns** and **exponential** relationships, although more complex forms of non-linearity are also possible. - The example to the left shows a clear diminishing returns between life expectancy and GDP per capita. --- ## Non-linearity can be hard to detect <img src="module6_slides_model_complications_files/figure-html/non-linear-movies-1.png" width="864" style="display: block; margin: auto;" /> --- ## Two techniques for detecting non-linearity -- .pull-left[ ### Non-linear smoothing <img src="module6_slides_model_complications_files/figure-html/smooth-example-1.png" width="504" style="display: block; margin: auto;" /> ] -- .pull-right[ ### Diagnostic residual plot <img src="module6_slides_model_complications_files/figure-html/resid-fitted-example-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Non-linear smoothing - A smoothing function uses the values of the `\(y\)` for the closest neighbors for a given observation of `\(x\)` to calculate a smoothed value of `\(y\)` that will reduce extreme values. - Smoothing functions vary by - What function is used to calculate the smoothed values (e.g. mean, median) - The span of how many neighbors are considered. Larger spans will lead to smoother lines. -- ### Median smoothing box office returns for *Rush Hour 3* .left-column[  ] .right-column[ <table> <thead> <tr> <th style="text-align:left;"> Movie </th> <th style="text-align:right;"> Tomato Rating </th> <th style="text-align:right;"> Box Office Returns </th> <th style="text-align:right;"> Smoothed Returns </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Awake </td> <td style="text-align:right;"> 4.2 </td> <td style="text-align:right;"> 14.3 </td> <td style="text-align:right;"> - </td> </tr> <tr> <td style="text-align:left;"> Ghost Rider </td> <td style="text-align:right;"> 4.2 </td> <td style="text-align:right;"> 115.8 </td> <td style="text-align:right;"> - </td> </tr> <tr> <td style="text-align:left;"> Rush Hour 3 </td> <td style="text-align:right;"> 4.2 </td> <td style="text-align:right;"> 140.1 </td> <td style="text-align:right;"> 32.8 </td> </tr> <tr> <td style="text-align:left;"> Balls of Fury </td> <td style="text-align:right;"> 4.2 </td> <td style="text-align:right;"> 32.8 </td> <td style="text-align:right;"> - </td> </tr> <tr> <td style="text-align:left;"> Nobel Son </td> <td style="text-align:right;"> 4.2 </td> <td style="text-align:right;"> 0.3 </td> <td style="text-align:right;"> - </td> </tr> </tbody> </table> ] --- ## Applying a median smoother <img src="module6_slides_model_complications_files/figure-html/median-smoothing-1.png" width="864" style="display: block; margin: auto;" /> --- ## The LOESS smoother .pull-left[ The LOESS (locally estimated scatterplot smoothing) estimates a smoothed value from a regression model of the focal point and neighbors. This model includes polynomial terms and weights observations to have more influence when close to the focal point. ### Use `geom_smooth` ```r ggplot(movies, aes(x=TomatoRating, y=BoxOffice))+ geom_jitter(alpha=0.2)+ * geom_smooth(method="loess", se=FALSE)+ scale_y_continuous(labels = scales::dollar)+ labs(x="Rotten Tomatoes Rating", y="Box Office Returns (millions USD)") ``` ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-10-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## <i class="fas fa-user-ninja "></i> Adjusting span .pull-left[ ```r ggplot(subset(gapminder, year==2007), aes(x=gdpPercap, y=lifeExp))+ geom_point(alpha=0.7)+ geom_smooth(method="loess", span=1, * se=FALSE, color="green")+ geom_smooth(method="loess", span=0.75, * se=FALSE, color="red")+ geom_smooth(method="loess", span=0.25, * se=FALSE, color="blue")+ labs(x="GDP per capita", y="Life expectancy at birth")+ scale_x_continuous(labels=scales::dollar) ``` The default `span` is 0.75 which is 75% of observations. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-11-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## ⚠️ LOESS with large `n` will 💀 your 💻 .pull-left[ ### Use a GAM instead Generalized Additive Models (GAM) are another way to create non-linear smoothing that is less computational intensive that LOESS but with similar results. ```r ggplot(subset(gapminder, year==2007), aes(x=gdpPercap, y=lifeExp))+ geom_point(alpha=0.7)+ geom_smooth(method="loess", se=FALSE, color="blue")+ * geom_smooth(method="gam", * formula = y ~ s(x, bs = "cs"), * se=FALSE, color="red")+ labs(x="GDP per capita", y="Life expectancy at birth")+ scale_x_continuous(labels=scales::dollar) ``` ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-12-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Or let `geom_smooth` figure out best smoother .pull-left[ ```r ggplot(earnings, aes(x=age, y=wages))+ geom_jitter(alpha=0.01, width=1)+ geom_smooth(method="lm", se=FALSE, color="blue")+ * geom_smooth(se=FALSE, color="red")+ scale_y_continuous(labels = scales::dollar)+ labs(x="age", y="hourly wages")+ theme_bw() ``` - For datasets over 1000 observations, `geom_smooth` will use GAM and otherwise defaults to LOESS. - Don't forget you can also specify a linear fit with `method="lm"`. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-13-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Residual plots .pull-left[ - A scatterplot of the residuals vs. the fitted values from a model can also be useful for detecting non-linearity. - If the relationship is linear, then we should expect to see no sign of a relationship in this plot. Drawing a smoothed line can be useful for this diagnosis. - Residual plots can also help to detect **heteroskedasticity** which we will talk about later. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-14-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Using `broom` and `augment` to get model data ```r library(broom) model <- lm(lifeExp~gdpPercap, data=subset(gapminder, year==2007)) augment(model) ``` ``` ## # A tibble: 142 x 9 ## lifeExp gdpPercap .fitted .se.fit .resid .hat .sigma .cooksd .std.resid ## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 43.8 975. 60.2 0.973 -16.4 0.0120 8.82 0.0207 -1.85 ## 2 76.4 5937. 63.3 0.818 13.1 0.00846 8.86 0.00928 1.48 ## 3 72.3 6223. 63.5 0.812 8.77 0.00832 8.90 0.00411 0.990 ## 4 42.7 4797. 62.6 0.848 -19.9 0.00907 8.77 0.0231 -2.25 ## 5 75.3 12779. 67.7 0.750 7.61 0.00709 8.91 0.00263 0.858 ## 6 81.2 34435. 81.5 1.52 -0.271 0.0292 8.93 0.0000144 -0.0309 ## 7 79.8 36126. 82.6 1.61 -2.75 0.0327 8.93 0.00167 -0.315 ## 8 75.6 29796. 78.5 1.29 -2.91 0.0211 8.93 0.00118 -0.331 ## 9 64.1 1391. 60.5 0.958 3.61 0.0116 8.93 0.000975 0.408 ## 10 79.4 33693. 81.0 1.48 -1.59 0.0278 8.93 0.000471 -0.181 ## # … with 132 more rows ``` --- ## Creating a residual plot .pull-left[ ```r ggplot(augment(model), aes(x=.fitted, y=.resid))+ geom_point()+ geom_hline(yintercept = 0, linetype=2)+ geom_smooth(se=FALSE)+ labs(x="fitted values of life expectancy", y="model residuals") ``` ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-15-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Its non-linear, so now what? -- .pull-left[ ### Transform variables <img src="module6_slides_model_complications_files/figure-html/transform-example-1.png" width="504" style="display: block; margin: auto;" /> ] -- .pull-right[ ### Polynomial terms <img src="module6_slides_model_complications_files/figure-html/polynomial-example-1.png" width="504" style="display: block; margin: auto;" /> ] -- .center[ ### Create splines <img src="module6_slides_model_complications_files/figure-html/spline-example-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Transforming variables .left-column[  ] .right-column[ A transformation is a mathematical function that changes the value of a quantitative variable. There are many transformations that one could apply, but we will focus on one - the **log** transformation. This is the most common transformation used in the social sciences. Transformations are popular because they can solve multiple problems: - A transformation can make a non-linear relationship look linear. - A transformation can make a skewed distribution more symmetric. - A transformation can reduce the impact of extreme outliers. ] --- ## Plotting the log transformation .pull-left[ ```r ggplot(subset(gapminder, year==2007), aes(x=gdpPercap, y=lifeExp))+ geom_point(alpha=0.7)+ geom_smooth(method="lm", se=FALSE)+ * scale_x_log10(labels=scales::dollar)+ labs(x="GDP per capita (log scale)", y="Life expectancy at birth") ``` - The scale of the independent variable is now multiplicative - The relationship looks more linear now, but what does it mean? ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-16-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## The natural log .left-column[  ] .right-column[ Although `ggplot` uses log with a base 10, we usually use the *natural log* transformation in practice. Both transformations have the same effect on the relationship, but the natural log provides results that are easier to interpret. In R, it is easy to take the natural log of a number by just using the `log` command. Any positive number can be logged. ```r log(5) ``` ``` ## [1] 1.609438 ``` The natural log of 5 is 1.609, but what does this mean? The natural log of any number is the power that you would have to raise the constant `\(e\)` to in order to get the original number back. In R, we can calculate `\(e^x\)` with `exp(x)`: ```r exp(log(5)) ``` ``` ## [1] 5 ``` ] --- ## The log transformation makes multiplicative relationships additive .left-column[  ] .right-column[ The key feature of the log transformation is that it makes multiplicative relationships additive. `$$log(x*y)=log(x)+log(y)$$` ```r log(5*4) ``` ``` ## [1] 2.995732 ``` ```r log(5)+log(4) ``` ``` ## [1] 2.995732 ``` We can use this feature to model relative (percent) change rather than absolute change in our models. ] --- ## Logging box office returns gives us a linear fit <img src="module6_slides_model_complications_files/figure-html/log-scale-movies-1.png" width="864" style="display: block; margin: auto;" /> --- ## What does it mean for the model? To fit the model, we can just use the log transformation directly in the formula of our `lm` command: ```r model <- lm(log(BoxOffice)~TomatoRating, data=movies) coef(model) ``` ``` ## (Intercept) TomatoRating ## 0.9727622 0.2406026 ``` `$$\log(\hat{returns}_i)=0.973+0.241(rating_i)$$` How do we interpret the slope? -- - A one point increase in rating is associated with a 0.241 increase in ... the log of box office returns? 😕 .center[ ### What does it mean? ] --- ## Converting to the original scale To get back to the original scale of box office returns for the dependent variable, we need to exponentiate both side side of the regression equation by `\(e\)`: -- `$$e^{\log(\hat{returns}_i)}=e^{0.973+0.241(rating_i)}$$` -- On the left hand side, I will get back predicted box office returns by the definition of logs. On the right hand side, I can apply some of the mathematical properties of logarithms and powers. -- `$$\hat{returns}_i=(e^{0.973})(e^{0.241})^{rating_i}$$` -- ```r exp(0.973) ``` ``` ## [1] 2.64587 ``` ```r exp(0.241) ``` ``` ## [1] 1.272521 ``` I now have: `$$\hat{returns}_i=(2.65)(1.27)^{rating_i}$$` --- ## A multiplicative relationship `$$\hat{returns}_i=(2.65)(1.27)^{rating_i}$$` Lets calculate predicted box office returns for Tomato Ratings of 0, 1, and 2. -- `\(\hat{returns}_i=(2.65)(1.27)^{0}=2.65\)` -- `\(\hat{returns}_i=(2.65)(1.27)^{1}=2.65(1.27)\)` -- `\(\hat{returns}_i=(2.65)(1.27)^{2}=2.65(1.27)(1.27)\)` -- - For each one unit increase in the independent variable, you multiply the previous predicted value by 1.27 to get the new predicted value. Therefore the predicted value increases by 27%. -- - The model predicts that movies with a zero Tomato rating make 2.65 million dollars, on average. Every one point increase in the Tomato rating is associated with a 27% increase in box office returns, on average. --- ## General form and interpretation `$$\log(\hat{y}_i)=b_0+b_1(x_i)$$` - You must apply the `exp` command to your intercept and slopes in order to interpret them. -- - The model predicts that the mean of `\(y\)` when `\(x\)` is zero will be `\(e^{b_0}\)`. -- - The model predicts that each one unit increase in `\(x\)` is associated with a multiplicative change of `\(e^{b_1}\)` in `\(y\)`. It is often easiest to express this in percentage terms. -- - An **absolute** change in `\(x\)` is associated with a **relative** change in `\(y\)`. --- ## Interpret these numbers ```r coef(lm(log(BoxOffice)~relevel(Rating, "PG"), data=movies)) ``` ``` ## (Intercept) relevel(Rating, "PG")G ## 3.2204949 0.4965918 ## relevel(Rating, "PG")PG-13 relevel(Rating, "PG")R ## -0.2351909 -1.9317768 ``` `$$\log(\hat{returns}_i)=3.22+0.50(G_i)-0.24(PG13_i)-1.93(R_i)$$` -- - `\(e^{3.22}=25\)`: PG-rated movies make 25 million dollars on average. -- - `\(e^{0.5}=1.64\)`: G-rated movies make 64% more than PG-rated movies, on average. -- - `\(e^{-0.24}=0.79\)`: PG-13 rated movies make 79% as much as (or 21% less than) PG-rated movies, on average. -- - `\(e^{-1.93}=0.14\)`: R-rated movies make 14% as much as (or 86% less than) PG-rated movies, on average. --- ## Approximating small effects When `\(x\)` is small (say `\(x<0.2\)`), then `\(e^x\approx1+x\)`. We can use this fact to roughly approximate coefficients/slopes as percent change when they are small. -- ```r model <- lm(log(BoxOffice)~Runtime, data=movies) coef(model) ``` ``` ## (Intercept) Runtime ## -1.96019099 0.04025076 ``` ```r exp(coef(model)) ``` ``` ## (Intercept) Runtime ## 0.1408315 1.0410718 ``` -- - If we do the full exponentiating, we can see that a one minute increase in runtime is associated with a 4.1% increase in box office returns. -- - The actual percentage increase is very close to what we got for the slope of the non-exponentiated slope (0.0402). So, you can often get a ballpark estimate without having to exponentiate. --- ## Logging the independent variable .pull-left[ Lets return to the relationship between GDP per capita and life expectancy that fit well as a linear relationship when we logged GDP per capita. Lets run the model: ```r model <- lm(lifeExp~log(gdpPercap), data=subset(gapminder, year==2007)) round(coef(model), 5) ``` ``` ## (Intercept) log(gdpPercap) ## 4.94961 7.20280 ``` How do we interpret these results? This case requires something different than the case where we logged the dependent variable. ] -- .pull-right[ Our basic model for life expectancy by GDP per capita is: `$$\hat{y}_i=4.9+7.2\log{x_i}$$` What is the predicted value of life expectancy at $1 GDP? `$$\hat{y}_i=4.9+7.2\log{1} = 4.9+7.2 * 0=4.9$$` What happens when we increase GDP per capita by 1% (from 1 to 1.01)? `$$\hat{y}_i=4.9+7.2\log{1.01} = 4.9+7.2 * 0.01=4.9+0.072$$` Predicted life expectancy increases by 0.072 years. ] --- ## General form and interpretation `$$\hat{y}_i=b_0+b_1\log(x_i)$$` -- - A one percent increase in `\(x\)` is associated with a `\(b_1/100\)` unit change in `\(y\)`, on average. -- - A **relative** change in `\(x\)` is associated with an **absolute** change in `\(y\)`. -- - `\(exp(b_0)\)` gives the predicted value of `\(y\)` when `\(x\)` equals one. -- - Keep in mind that the `\(log(0)\)` is negative infinity so you cannot predict the value of `\(y\)` when `\(x=0\)`. --- ## Logging both variables <img src="module6_slides_model_complications_files/figure-html/transform-wages-1.png" width="864" style="display: block; margin: auto;" /> --- ## The elasticity model ```r model <- lm(log(wages)~log(age), data=earnings) coef(model) ``` ``` ## (Intercept) log(age) ## 1.1558386 0.5055849 ``` -- - This is actually the easiest model to interpret. We can interpret the slope directly as the percent change in `\(y\)` for a one percent increase in `\(x\)`. -- - Calculating the percent change in one variable by percent change in another is what economists call an **elasticity** so this model is often called an **elasticity** model. -- - The model predicts that a one percent increase in age is associated with a 0.51% increase in wages, on average. --- ## A cheat sheet | Which variable logged | Non-linear shape | Change in x | Change in y | Interpret `\(\beta_1\)` | |-----------------------|--------------------|-------------|-------------|---------------------| | Independent variable | diminishing returns| relative | absolute | `\(\beta_1/100\)` | | Dependent variable | exponential | absolute | relative | `\(e^{\beta_1}\)` | | Both variables | both types | relative | relative | `\(\beta_1\)` | --- ## When `\(x<=0\)`, logging is bad .pull-left[ ### Log problems `$$log(0)=-\infty$$` `$$log(x)\text{ is undefined when }x<0$$` ### Try the square/cube root - The square root transformation has a similar effect to the log transformation but can include zero values. - The cube root transformation can also include negative values. - The downside of square/cube root transformations is that values are not easy to interpret. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/sqrt-transform-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Polynomial models .pull-left[ ### Polynomial expression A **polynomial** expression is one that adds together terms involving multiple powers of a single variable. if we include a squared value of `\(x\)` we get the classic formula for a parabola: `$$y=a+bx+cx^2$$` <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-24-1.png" width="504" style="display: block; margin: auto;" /> ] -- .pull-right[ ### A polynomial model We can fit such a parabola in a linear model by including a new variable that is simply the square of the original variable: `$$\hat{y}_i=\beta_0+\beta_1x_i+\beta_2x_i^2$$` ```r model <- lm(wages~I(age-40)+I((age-40)^2), data=earnings) coef(model) ``` ``` ## (Intercept) I(age - 40) I((age - 40)^2) ## 26.92134254 0.31711167 -0.01783204 ``` Our model is: `$$\hat{y}_i=26.9+0.3171(x_i-40)-0.0178(x_i-40)^2$$` How do we interpret the results? ] --- ## Calculate the marginal effect With polynomial terms, the marginal effect of `\(x\)` is more complicated because our `\(x\)` shows up in more than one place in the equation: `$$y_i=\beta_0+\beta_1x_i+\beta_2x_i^2+\epsilon_i$$` -- By calculus, we can mathemagically get the marginal effect for this model as: `$$\frac{\partial y}{\partial x}=\beta_1+\beta_2x$$` -- So, for our case: `$$\hat{y}_i=26.9+0.3171(x_i-40)-0.0178(x_i-40)^2$$` the marginal effect of age on wages is given by: `$$0.3171+2*-0.0178(x-40)=0.3171-0.0356(x-40)$$` - At age 40 (the zero value), a one year increase in age is associated with a salary increase of $0.32, on average. - For every year over 40, this increase is smaller by $0.0356. For every age younger than 40, this increase is larger by $0.0356. --- ## Finding the inflection point .pull-left[ If the effect of age on wages goes down by $0.0356 for every year over 40, then at some point the positive effect of age on wages will become negative. We can figure out the value of age at this **inflection point** by setting the effect to zero and solving for x. In general, this will give us: `$$\beta_1/(-2*\beta_2)$$` In our case, we get: `$$0.3171/(-2*-0.0178)=0.3171/0.0356=8.91$$` So the model predicts that the effect of age on wages will shift from positive to negative at age 48.91. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/inflection-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Plotting a polynomial fit .pull-left[ ```r ggplot(earnings, aes(x=age, y=wages))+ geom_jitter(alpha=0.01, width=1)+ geom_smooth(se=FALSE, * method="lm", * formula=y~x+I(x^2))+ labs(x="age", y="hourly wages") ``` in `geom_smooth`, you can specify a formula for the `lm` method that includes a squared term. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-25-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Higher order terms gives you more wiggle .pull-left[ ```r ggplot(subset(gapminder, year==2007), aes(x=gdpPercap, y=lifeExp))+ geom_point()+ geom_smooth(se=FALSE, method="lm", formula=y~x+I(x^2), color="blue")+ geom_smooth(se=FALSE, method="lm", formula=y~x+I(x^2)+I(x^3), color="red")+ geom_smooth(se=FALSE, method="lm", formula=y~x+I(x^2)+I(x^3)+I(x^4), color="green")+ labs(x="GDP per capita", y="life expectancy") ``` ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-26-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Spline models .pull-left[ <img src="module6_slides_model_complications_files/figure-html/spline-wages-1.png" width="504" style="display: block; margin: auto;" /> ] .pull-right[ - The basic idea of a spline model is to allow the slope of the relationship between `\(x\)` and `\(y\)` to be different at different cutpoints or "hinge" values of `\(x\)`. - These cutpoints create different linear segments where the effect of x on y is different. - We will look at the case of one hinge value which gives us an overall slope that looks like a "broken arrow." ] --- ## Creating the spline variable The relationship between age and wages suggests that the relationship shifts considerably around age 35. To model this we create a spline variable like so: `$$spline_i=\begin{cases} age-35 & \text{if age>35}\\ 0 & \text{otherwise} \end{cases}$$` We can then add this variable to our model: ```r earnings$age.spline <- ifelse(earnings$age<35, 0, earnings$age-35) model <- lm(wages~age+age.spline, data=earnings) coef(model) ``` ``` ## (Intercept) age age.spline ## -6.0393027 0.9472390 -0.9549087 ``` How do we interpret the results? --- ## Interpreting results in spline model .pull-left[ Up to age 35, the spline term is zero, so our model is given by: `$$\hat{wages}_i=-6.04+0.9472(age_i)$$` The model predicts that for individuals age 35 and under, a one year increase in age is associated with a $0.9472 increase in wages, on average. After age 35, the spline variable increases by one every time age increases by one, so the marginal effect of age is given by: `$$0.9472-09549=-0.0077$$` The model predicts that for individuals over age 35, a one year increase in age is associated with a $0.0077 reduction in wages, on average. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/spline-interpret-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## <i class="fas fa-user-ninja "></i> Plotting the spline model .pull-left[ We cannot use `geom_smooth` to show the fit of this spline model. However, with some additional effort, we can plot the effect on a graph. 1. Calculate the predicted value of wages for a range of age values based on the model using the `predict` command. 2. Feed in these predicted values as a dataset to the `geom_line` function in our `ggplot` graph. ] -- .pull-right[ ### The `predict` command The `predict` command takes two important arguments: 1. The model object for which we want predicted values of the dependent variable. 2. A new dataset that contains all the same independent variables that are in the model. ```r model <- lm(wages~age+age.spline, data=earnings) pre_df <- data.frame(age=18:65) pre_df$age.spline <- ifelse(pre_df$age<35, 0, pre_df$age-35) pre_df$wages <- predict(model, newdata=pre_df) head(pre_df, n=3) ``` ``` ## age age.spline wages ## 1 18 0 11.01100 ## 2 19 0 11.95824 ## 3 20 0 12.90548 ``` ] --- ## Adding the spline fit to ggplot .pull-left[ ```r ggplot(earnings, aes(x=age, y=wages))+ geom_jitter(alpha=0.01, width = 1)+ * geom_line(data=predict_df, color="blue", size=1.5) ``` To add the spline fit, we just add a `geom_line` command and specify our `pre_df` dataset through the `data` argument so that it uses the predicted values rather than the actual `earnings` dataset to graph the line. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-27-1.png" width="504" style="display: block; margin: auto;" /> ] --- class: inverse, center, middle background-image: url(images/bence-balla-schottner-o5ttfkZU9_Q-unsplash.jpg) background-size: cover name: iid # The IID Violation and Robust Standard Errors --- ## Linear model as data-generating process `$$y_i=\underbrace{\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\ldots+\beta_px_{ip}}_{\hat{y}_i}+\epsilon_i$$` -- - To get values of `\(y_i\)`, you feed in values of `\(x_i\)` to the structural component and get back out a predicted `\(\hat{y_i}\)` value. -- - To get the stochastic (random) part, you then reach into some distribution to grab a random value of `\(\epsilon_i\)` that you add to your predicted value to get an actual value of `\(y_i\)`. -- - What distribution are you reaching into when you grab `\(\epsilon_i\)`? The linear model framework assumes two things about this distribution: -- - **Independence**: The number you pull out each time doesn't depend on other numbers that you pull out. This is most commonly violated by **autocorrelation** or the clustering of **repeated observations**. -- - **Identical distribution**: You reach into the same distribution for all observations. This is most commonly violated by **heteroscedasticity** which means non-constant variance in the residuals. -- - Together these assumptions give us the **IID** assumption of the linear model: the error terms are **independent and identically distributed**. --- ## Violations of independence -- .pull-left[ ### Serial autocorrelation <img src="module6_slides_model_complications_files/figure-html/longley-1.png" width="504" style="display: block; margin: auto;" /> In time series data, sequential observations in time are likely to either be highly positively or negatively correlated. In this case, when one year has a higher or lower than expected value, the next and previous years are also likely to have a higher or lower than expected value. ] -- .pull-right[ ### Repeated observations .center[] When observations are drawn repeatedly from a sample of some larger units (i.e. a multilevel structure), then observations within the same unit are likely to vary from predicted values in the same way. For example, students in the same classroom might tend to have either lower or higher test scores than predicted by the model due to some unobserved feature of that classroom. ] --- ## Serial autocorrelation example .pull-left[ ```r model <- lm(Employed~GNP, data=longley) temp <- augment(model)$.resid temp <- data.frame(x=temp[1:15], y=temp[2:16]) ggplot(temp, aes(x=x, y=y))+ geom_point()+ geom_smooth(method="lm", se=FALSE)+ geom_hline(yintercept = 0, linetype=2)+ labs(x="residuals from 1947-1961", y="residuals from 1948-1962")+ annotate("text",0.25, 0.5, label="r=0.16") ``` As an example of serial autocorrelation, I will use the `longley` time series dataset in R to fit the following model predicting the number of people employed by GNP from 1947 to 1962 (n=16): I can then plot the residuals values from years 1947 to 1961 by the residual values for years 1948 to 1962. The positive correlation in the residuals here suggests serial autocorrelation. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/unnamed-chunk-28-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Heteroscedasticity .pull-left[ .center[] - Heteroscedasticity means that the variance of the residuals is not constant but depends on the values of `\(x_i\)`, and therefore, implicitly, `\(\hat{y}_i\)`. - A classic example of heteroscedasticity is when the variance of the residuals increases with larger values of `\(\hat{y}_i\)` giving you a cone shape in a residual by fitted value plot. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/heteroscedasticity-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Correcting for iid violations Violating the iid. assumption does not bias your results, but it will lead to inefficient estimates and poorly estimated standard errors. There are a number of potential solutions to the iid problem. These include: -- - **Transformations** (particularly the log transformation) can often solve the problem of heteroscedasticity. -- - **Weighted least squares** models can correct for iid when the nature of the violation is understood. -- - **Robust standard errors** can be used as a crude brute-force solution when the nature of the violation is not well understood. I would recommend that this only be done for diagnostic reasons. -- - In general, the best approach is to re-think your model. If you have an iid violation then you are probably not applying the best type of model to the problem at hand. --- ## Fixing heteroscedasticity with a transformation .pull-left[ <img src="module6_slides_model_complications_files/figure-html/heteroscedasticity-original-1.png" width="504" style="display: block; margin: auto;" /> ] -- .pull-right[ <img src="module6_slides_model_complications_files/figure-html/heteroscedasticity-log-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Using Weighted Least Squares The weighted least squares technique uses a weighting matrix `\(\mathbf{W}\)` in its calculation of regression slopes like so: `$$\mathbf{\beta}=(\mathbf{X'W^{-1}X})^{-1}\mathbf{X'W^{-1}y}$$` `$$SE_{\beta}=\sqrt{\sigma^{2}(\mathbf{X'W^{-1}X})^{-1}}$$` The exact form of this weighting matrix depends on the nature of the iid violation, but in general it is used to represent the covariance between residuals. Values in the diagonal cells adjust for heteroscedasticity and values in other cells adjust for autocorrelation. This technique is used routinely in time series analysis to adjust for serial autocorrelation. --- ## GLS Example We can use the `gls` command in the `nmle` package to adjust for serial autocorrelation in the prior model.We will assume that the autocorrelation follows an "AR1" pattern in which each subsequent residual is only correlated with its immediate predecessor (AR1 stands for auto-regressive 1, where 1 indicates the lag). ```r summary(lm(Employed~GNP+Population, data=longley))$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 88.93879831 13.78502699 6.451841 2.160219e-05 ## GNP 0.06317244 0.01064721 5.933238 4.958268e-05 ## Population -0.40974292 0.15213679 -2.693253 1.842949e-02 ``` ```r library(nlme) model.ar1 <- gls(Employed~GNP+Population, correlation=corAR1(form=~Year),data=longley) summary(model.ar1)$tTable ``` ``` ## Value Std.Error t-value p-value ## (Intercept) 101.85813305 14.1989324 7.173647 7.222049e-06 ## GNP 0.07207088 0.0106057 6.795485 1.271171e-05 ## Population -0.54851350 0.1541297 -3.558778 3.497130e-03 ``` --- ## Robust Standard Errors .pull-left[ We won't delve into the math behind the robust standard error, but the general idea is that robust standard errors will give you "correct" standard errors even when the model is mis-specified due to issues such a non-linearity, heteroscedasticity, and autocorrelation. Robust standard errors can be estimated in R using the `sandwich` and `lmtest` packages, and specifically with the `coeftest` command. Within this command, it is possible to specify different types of robust standard errors, but we will use the "HC1" version which is equivalent to the robust standard errors produced in Stata by default. ] -- .pull-right[ ```r library(sandwich) library(lmtest) model <- lm(BoxOffice~TomatoRating, data=movies) summary(model)$coef[,1:2] ``` ``` ## Estimate Std. Error ## (Intercept) -16.71941 5.1103643 ## TomatoRating 11.43015 0.9140331 ``` ```r coeftest(model, vcov=vcovHC(model, "HC1"))[,1:2] ``` ``` ## Estimate Std. Error ## (Intercept) -16.71941 5.257507 ## TomatoRating 11.43015 1.097638 ``` Note that the estimates are the same, but the robust standard errors are considerably larger. That difference in magnitude is telling us that our basic regression model is problematic. In this case, we already know that the problem is heteroscedasticity. ] --- ## ⚠️ Robust standard errors diagnose problems that they do not fix! ```r model <- lm(log(BoxOffice)~TomatoRating, data=movies) summary(model)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.9727622 0.20091999 4.841540 1.365329e-06 ## TomatoRating 0.2406026 0.03593629 6.695254 2.638277e-11 ``` ```r coeftest(model, vcov=vcovHC(model, "HC1")) ``` ``` ## ## t test of coefficients: ## ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 0.972762 0.186379 5.2193 1.941e-07 *** ## TomatoRating 0.240603 0.032115 7.4919 9.302e-14 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` When the dependent variable is logged to remove heteroscedasticity, the difference between robust and regular standard errors goes away. --- class: inverse, center, middle background-image: url(images/cyril-saulnier-TsVN31Dzyv4-unsplash.jpg) background-size: cover name: sample-design # Sample Design and Weighting --- ## The reality of survey sampling -- .pull-left[ ### The simple random sample .center[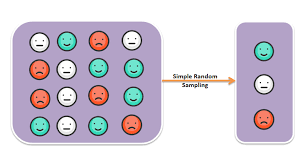] - We have treated all of our sample data as if it was collected using simple random sampling (SRS). - In an SRS of size `\(n\)`, every possible combination of `\(n\)` observations from the population has an equally likely chance of being drawn. - In an SRS, every statistic (e.g. mean, proportions, linear slopes) should be representative of the population, except for random sampling bias. ] -- .pull-right[ ### Reality .center[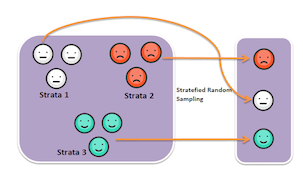] - In practice, large-scale surveys never use SRS for pragmatic and design reasons. - For correct statistical inference, we typically need to make adjustments for sample design. - The primary ways in which sample design affects estimation are**clustering**, **stratification**, and **weighting**. ] --- ## Cluster/Multistage sampling .left-column[ 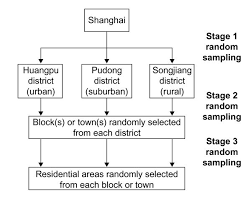 ] .right-column[ Potential observations are aggregated into larger groupings identified as the Primary Sampling Unit (PSU) and then we sample some of these PSUs before sampling individual observations within the sampled PSUs. - Cluster sampling is about efficiency and cost. Clusters are typically defined geographically, which then minimizes the cost of sampling individual observations. - If PSUs are sampled with probabilities proportional to cluster size, then every unit in the population has an equal likelihood of being selected. In this case, summary statistics on the sample should be representative. - When the variable of interest is distributed differently across clusters, the sampling variability will be higher than an SRS even if every observation has an equally likely chance of being drawn. ] --- ## What percent of the US population is Mormon? .pull-left[ The General Social Survey uses Metropolitan Statistical Areas (MSA) and clusters of non-metropolitan counties as the PSU. In each year, it draws a sample of these areas and then samples respondents within each sampled PSU. Because Mormons are heavily concentrated in certain places, percent Mormon in the GSS varies substantially from year to year by whether certain PSUs were sampled or not. Red band indicates expected 95% interval for sampling variability without clustering, assuming the average percent across all years (dotted line). ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/mormons-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Stratification Stratification in sampling operates in a manner somewhat similar to cluster sampling except that once the observations are aggregated into strata by some characteristic (e.g. income, race, age), observations are sampled from *every* stratum. -- - Stratification is typically done to ensure that various sub-populations are present within the sample. -- - Different strata may be sampled with different probabilities. The most common approach is to take an **oversample** of a small group in order to ensure that effective comparisons can be made between that group and other groups. -- - In practice, stratification is often done by first screening potential respondents for stratum characteristics. -- - If strata are sampled with different probabilities, then summary statistics for the full sample will not be representative without weight adjustments. -- - Unlike clustering, greater similarity on a characteristic of interest within strata can actually **reduce** the sampling variability for that characteristic relative to an SRS. --- ## Weighting -- - Numerous factors can lead to a sample being unrepresentative such as sampling strata with different probabilities, differential non-response rates, and a lack of fit between sample frame and population. -- - **Sampling weights** allow researchers to correct summary statistics from the sample so that they are representative of the population. -- - The sampling weight for an observation should be `\(1/p_i\)` where `\(p_i\)` is the probability of being sampled. The sampling weight indicates the number of observations in the population that observation `\(i\)` in the sample represents. -- - Calculating sampling weights can be quite complex. In some cases, researchers may know `\(p_i\)` from the study design. In other cases, researchers may create **post-stratification** weights by comparing the sample to some other data source (e.g. census, school records) for a set of demographic characteristics and applying weights to make the sample align with the other data source. -- - When sampling weights are present in a dataset, they must be used to generate statistics representative of the population. -- - Variation in sampling weights will increase sampling variability above and beyond that expected for an SRS. --- ## Weights in Sneetchville .left-column[  ] .right-column[ In Sneetchville, there are 3 star-bellied and 7 regular sneetches. Lets say I take a stratified sample of two star-bellied and two regular sneetches. Here is the population data: ```r sneetches <- data.frame(type=factor(c(rep("Star-bellied", 3),rep("Regular",7))), income=c(3,7,6,2,1,4,5,0,2,1)) sneetches$prob <- c(rep(2/3,3),rep(2/7,7)) sneetches ``` ``` ## type income prob ## 1 Star-bellied 3 0.6666667 ## 2 Star-bellied 7 0.6666667 ## 3 Star-bellied 6 0.6666667 ## 4 Regular 2 0.2857143 ## 5 Regular 1 0.2857143 ## 6 Regular 4 0.2857143 ## 7 Regular 5 0.2857143 ## 8 Regular 0 0.2857143 ## 9 Regular 2 0.2857143 ## 10 Regular 1 0.2857143 ``` ] --- ## Weights in Sneetchville ```r sneetch.sample <- sneetches[c(sample(1:3,2),sample(4:10,2)),] sneetch.sample$weight <- 1/sneetch.sample$prob sneetch.sample ``` ``` ## type income prob weight ## 2 Star-bellied 7 0.6666667 1.5 ## 3 Star-bellied 6 0.6666667 1.5 ## 7 Regular 5 0.2857143 3.5 ## 6 Regular 4 0.2857143 3.5 ``` ```r x <- c(mean(sneetches$income), mean(sneetch.sample$income), weighted.mean(sneetch.sample$income, sneetch.sample$weight)) names(x) <- c("population","sample.unweighted","sample.weighted") x ``` ``` ## population sample.unweighted sample.weighted ## 3.1 5.5 5.1 ``` --- ## The consequences of survey design | Design issue | Representative? | Design effect (change to SE) | |:-----------------|:--------------------------------------------------------------------------------|:--------------------------------------------| | Clustering | Yes, if PSUs are proportionally drawn. Otherwise, weighting necessary. | Increases with difference between clusters. | | Stratification | Yes, if strata are sampled with same probability. Otherwise weighting necessary.| Decreases with homogeneity within strata | | Weights | Only if weights are applied. | Increases with the variance of the weights. | --- ## Correcting for survey design -- ### Basic Weighting R has syntax in many commands to apply sampling weights to get representative statistics (e.g. `weighted.mean`, the `weight` option in the `lm` command), but this approach will not correctly adjust standard errors for design effects. -- ### Robust standard errors Robust standard errors will correct standard errors for differential weights, but not for clustering and stratification design effects. -- ### Survey package The `survey` package in R will allow you to specify design and correctly adjust standard errors. --- ## Add Health survey design .pull-left[ 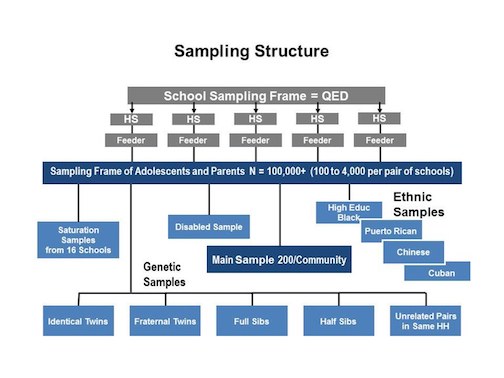 ] .pull-right[ - Schools were the primary PSU in a cluster sampling technique, but were also stratified by region, urbanicity, school type, ethnic mix, and size. - Students within schools were stratified by grade and sex and then sampled. - Several oversamples were conducted of ethnic groups and genetically related pairs of students, as well as saturated samples from 16 schools. - Post-stratification adjustments were made to sampling weights to account for region of the country. - The Add Health documentation indicates that the REGION variable should be applied as a stratification variable but this variable is not available in the public release data so we will focus on the design effects of weights and clustering. ] --- ## Sample design variables .pull-left[ ```r head(popularity[,c("cluster","sweight")]) ``` ``` ## cluster sweight ## 1 472 3663.6562 ## 2 472 4471.7647 ## 3 142 3325.3294 ## 4 142 271.3590 ## 5 145 3939.9402 ## 6 145 759.0284 ``` - `cluster` is a numeric id indicating the school the student was sampled from. - `sweight` is the inverse of the probability of being sampled. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/addhealthweight-dist-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Add Health example, naive approach Lets start with a model that makes no adjustment. ```r model.basic <- lm(indegree~nsports, data=popularity) summary(model.basic)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 3.9969128 0.07221383 55.34830 0.000000e+00 ## nsports 0.5044705 0.04282271 11.78044 1.462129e-31 ``` -- Because I am not using weights, this value will not be representative of all American high school students in 1994-95. I can add the weights provided by Add Health to make this representative: -- ```r model.weight <- update(model.basic, weight=sweight) summary(model.weight)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.0268786 0.07321643 54.99966 0.00000e+00 ## nsports 0.5330072 0.04347144 12.26109 5.23695e-34 ``` --- ## Add Health example, robust SE approach Adding weights to the `lm` command gives me representative values for the slope and intercept, but my SEs do not correct for the substantial variation in weights. I can correct for this design issue by using robust standard errors: -- ```r library(sandwich) library(lmtest) model.robust <- coeftest(model.weight, vcov=vcovHC(model.weight, "HC1")) model.robust ``` ``` ## ## t test of coefficients: ## ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.026879 0.080106 50.2692 < 2.2e-16 *** ## nsports 0.533007 0.063041 8.4549 < 2.2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` Estimates are identical but standard errors are slightly larger. However, this technique still does not correct for clustering. --- ## Add Health example, using `survey` library I can use the `svydesign` command in the `survey` package to correctly specify both the weights and the clustering in Add Health. The `ids` argument expects a variable name that identifies the clusters by id (in this case, the school id) and the `weight` argument expects a variable name for the weights used. ```r library(survey) popularity.svy <- svydesign(ids=~cluster, weight=~sweight, data=popularity) model.svy <- svyglm(indegree~nsports, design=popularity.svy) summary(model.svy)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.0268786 0.11483268 35.067357 6.996643e-62 ## nsports 0.5330072 0.07318866 7.282648 5.034252e-11 ``` Estimates are identical to the weighted regression model before, but standard errors have increased due to variation in weights and the cluster design effect. --- ## Comparison of methods .stargazer[ <table cellspacing="0" align="center" style="border: none;"> <caption align="top" style="margin-bottom:0.3em;">Linear models predicting number of friend nominations by sports played</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>unweighted </b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>weighted </b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>robust SE </b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>survey: weights </b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>survey: weights+cluster</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">3.997<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.027<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.027<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.027<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.027<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.072)</td> <td style="padding-right: 12px; border: none;">(0.073)</td> <td style="padding-right: 12px; border: none;">(0.080)</td> <td style="padding-right: 12px; border: none;">(0.080)</td> <td style="padding-right: 12px; border: none;">(0.115)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Number of sports</td> <td style="padding-right: 12px; border: none;">0.504<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.533<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.533<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.533<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.533<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.063)</td> <td style="padding-right: 12px; border: none;">(0.063)</td> <td style="padding-right: 12px; border: none;">(0.073)</td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.031</td> <td style="border-top: 1px solid black;">0.033</td> <td style="border-top: 1px solid black;"></td> <td style="border-top: 1px solid black;"></td> <td style="border-top: 1px solid black;"></td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">4397</td> <td style="border-bottom: 2px solid black;">4397</td> <td style="border-bottom: 2px solid black;"></td> <td style="border-bottom: 2px solid black;">4397</td> <td style="border-bottom: 2px solid black;">4397</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="7"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] -- - All models except the unweighted version produce the same estimates of slope and intercept based on the weights. -- - The survey weighted and robust SE models both produce the same standard errors. This is because both models account for the design effect of weight heterogeneity but not clustering. --- class: inverse, center, middle background-image: url(images/kyaw-tun-UvL9JrLkagM-unsplash.jpg) background-size: cover name: missing-data # Missing Data --- ## The reality of missing data Missing data exists in most real-world data sets. Therefore, its important to know how to handle missing data in order to know how to properly conduct an analysis. -- .pull-left[ ### Valid Skip  Valid skips most arise when a follow-up question is only asked of respondents who gave a certain response to the initial question. If you construct a variable correctly, valid skips should not be considered missing values. ] -- .pull-right[ ### Item non-response  Item non-response occurs when respondents fail to respond to a specific question. This may be because they don't know the correct response or they do not feel comfortable answering the question. ] --- ## Example of a valid skip The GSS uses three variables to determine respondents' religious affiliation. - `relig` asks for major religious affiliations such as Catholic, Protestant, Jewish, Muslim, etc. - **If and only if** respondents indicate they are Protestant, they are asked a follow up question recorded in `denom` which asks for their specific denomination. - `denom` only lists major Protestant denominations. If the respondent checks "other", their specific write-in response is recorded in a third variable titled `other`. -- ```r summary(relig[,c("relig","denom","other")]) ``` ``` ## relig denom other ## protestant:34597 other : 7491 pentecostal : 878 ## catholic :14532 baptist-dk which: 6078 mormon : 714 ## none : 6635 southern baptist: 3613 church of christ : 648 ## jewish : 1195 no denomination : 3101 christian : 538 ## other : 1025 united methodist: 2693 jehovah's witnesses: 397 ## (Other) : 1592 (Other) :11959 (Other) : 4224 ## NA's : 23 NA's :24664 NA's :52200 ``` -- There are a lot of missing values for `denom` and `other`, but these are all valid skips based on prior responses. The only true missing values in this set of variables are the 23 respondents who did not respond to the initial question on `relig`. --- ## Kinds of missingness .left-column[  ] .right-column[ ### Missing Completely at Random (MCAR) Every observation has the same probability of missingness and the missingness of a variable has no relationship to other observed or unobserved variables. If this is true, then removing observations with missing values will not bias results. ### Missing at Random (MAR) The different probabilities of missingness can be fully accounted for by other observed variables in the dataset. If this is true, then various techniques can be used to produce unbiased results by **imputing** values for the missing values. ### Not Missing at Random (NMAR) The different probabilities of missingness depend both on observed and unobserved variables. In this case, we cannot fully correct for bias that might result from missing data. ] ??? In practice, its impossible to distinguish perfectly between MAR and NMAR. If we use many predictors to impute a moderate number of missing values for a case then the MAR assumption is reasonable in the sense that the remaining bias will likely be minimal. --- ## Add Health income example As an example, I will use parental income from the Add Health data to predict popularity. Income is recorded in thousands of dollars, and I have top-coded the values to $200,000. Income is notorious as a variable that will typically have a high non-response rate. The Add Health data are no different: ```r summary(addhealth$parentinc) ``` ``` ## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's ## 0.00 23.00 40.00 45.37 60.00 200.00 1027 ``` ```r mean(is.na(addhealth$parentinc)) ``` ``` ## [1] 0.2335683 ``` Income is missing for 1027 cases which is roughly a quarter of the dataset. * Note that the `popularity` dataset will not work here as I already imputed missing values. Use the `addhealth` dataset instead. --- ## Two basic approaches to missing values -- .pull-left[  ### Remove cases - Case deletion is easy to implement. - Case deletion assumes MCAR. - Case deletion may result in a substantial reduction is sample size. ] -- .pull-right[ .center[] ###Impute cases - Imputation is more difficult to implement. - If done properly, imputation assumes MAR. - Imputation will not reduce sample size. ] --- ## Two kinds of case deletion -- .pull-left[ ### Complete-case analysis (listwise deletion) - All cases that have missing values on any of the variables that will be used at some point in the analysis are removed from the start. - This ensures that all results are based on the same sample. ] -- .pull-right[ ### Available-case analysis (pairwise deletion) - Cases are removed model by model or statistic by statistic when the variables used in that particular statistic or model have missing values. - This is by default what will happen in R across different `lm` models with different variables. - This approach allows the researcher to use more data, but different statistics and models will use different subsets of the full data which makes comparability problematic. ] --- ## Removing cases, Add Health example ### Available-case analysis This is what happens by default when you just run nested models in R ```r model1.avail <- lm(indegree~nsports, data=addhealth) model2.avail <- update(model1.avail, .~.+alcoholuse+smoker) model3.avail <- update(model2.avail, .~.+parentinc) ``` -- ### Complete-case analysis To do this, we need to use `na.omit` on the Add Health variables that will be in the most complex model to make sure all models work with the same subset. ```r addhealth.complete <- na.omit(addhealth[,c("indegree","nsports","alcoholuse","smoker","parentinc")]) model1.complete <- lm(indegree~nsports, data=addhealth.complete) model2.complete <- update(model1.complete, .~.+alcoholuse+smoker) model3.complete <- update(model2.complete, .~.+parentinc) ``` --- ## What is different? .pull-left[ .stargazer[ ### Available-case method <table cellspacing="0" align="center" style="border: none;"> <caption align="bottom" style="margin-top:0.3em;">Statistical models</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 1</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 2</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 3</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">4.00<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.85<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.39<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.07)</td> <td style="padding-right: 12px; border: none;">(0.08)</td> <td style="padding-right: 12px; border: none;">(0.12)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Number of sports</td> <td style="padding-right: 12px; border: none;">0.50<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.51<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.45<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.04)</td> <td style="padding-right: 12px; border: none;">(0.04)</td> <td style="padding-right: 12px; border: none;">(0.05)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Drinker</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.71<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.66<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.16)</td> <td style="padding-right: 12px; border: none;">(0.18)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Smoker</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.19</td> <td style="padding-right: 12px; border: none;">0.38<sup style="vertical-align: 0px;">*</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.16)</td> <td style="padding-right: 12px; border: none;">(0.19)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Parental income (1000s USD)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.01<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.00)</td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.03</td> <td style="border-top: 1px solid black;">0.04</td> <td style="border-top: 1px solid black;">0.05</td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">4397</td> <td style="border-bottom: 2px solid black;">4343</td> <td style="border-bottom: 2px solid black;">3332</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="5"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] ] .pull-right[ .stargazer[ ### Complete-case method <table cellspacing="0" align="center" style="border: none;"> <caption align="bottom" style="margin-top:0.3em;">Statistical models</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 1</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 2</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 3</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">4.05<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.88<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.39<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.08)</td> <td style="padding-right: 12px; border: none;">(0.09)</td> <td style="padding-right: 12px; border: none;">(0.12)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Number of sports</td> <td style="padding-right: 12px; border: none;">0.49<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.49<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.45<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.05)</td> <td style="padding-right: 12px; border: none;">(0.05)</td> <td style="padding-right: 12px; border: none;">(0.05)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Drinker</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.72<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.66<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.18)</td> <td style="padding-right: 12px; border: none;">(0.18)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Smoker</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.37<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">0.38<sup style="vertical-align: 0px;">*</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.19)</td> <td style="padding-right: 12px; border: none;">(0.19)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Parental income (1000s USD)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.01<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.00)</td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.03</td> <td style="border-top: 1px solid black;">0.04</td> <td style="border-top: 1px solid black;">0.05</td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">3332</td> <td style="border-bottom: 2px solid black;">3332</td> <td style="border-bottom: 2px solid black;">3332</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="5"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] ] -- ??? - Number of observations changes across models in available-case analysis. - The results for smoking in the complete-case analysis suggest that the difference from model 2 to model 3 in available-case analysis was driven by sample size change, not controlling for income. --- ## Imputation -- .pull-left[ ### Predictive or non-Predictive? - Imputations vary by whether or not they use other predictors in the data to assign imputed values to missing values. - Predictive imputation is done via a statistical model. - Predictive imputation moves the assumption from MAR to MCAR if the model is good. - Non-predictive imputation will systematically bias correlation between variables downward. ] -- .pull-right[ ### Randomness? - Imputations vary by whether or not they include a random component or are purely deterministic. - Deterministic imputation techniques will underestimate variance in the imputed variable and will therefore underestimate standard errors. - The use of randomization leads to another source of variation called **imputation variation** which can only be fully addressed through the technique of **multiple imputation**. ] --- ## Non-Predictive Imputation A very simple (and poor) technique would be just to substitute the mean for valid responses for all missing values. ```r addhealth$parentinc.meani <- ifelse(is.na(addhealth$parentinc), mean(addhealth$parentinc,na.rm=TRUE), addhealth$parentinc) ``` -- Another similar technique that allows for more randomness is just to sample a random valid response on the same variable for each missing value. ```r addhealth$parentinc.randi <- addhealth$parentinc addhealth$parentinc.randi[is.na(addhealth$parentinc)] <- sample(na.omit(addhealth$parentinc), sum(is.na(addhealth$parentinc))) ``` --- ## Mean and random imputation, Add Health .pull-left[ <img src="module6_slides_model_complications_files/figure-html/meanimpute_plot-1.png" width="504" style="display: block; margin: auto;" /> ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/randomimpute_plot-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Non-predictive imputation == Bad | Sample | `\(r\)` (indegree and income) | SD (income) | |:--------------------|--------------------------:|-------------------:| | Valid cases | 0.1320575 | 33.6984995| | Valid cases +mean imputed | 0.1167858 | 29.5006936| | Valid cases +random imputed | 0.1076341 | 33.8174488| - Both techniques will systematically underestimate correlation. - Mean imputation will underestimate the variance of the imputed variable. --- ## Quick and dirty method If I do a mean imputation, I can also add the boolean variable indicating missingness as a predictor in my model. When I do this, the effect of the imputed variable will be the same as if if I had thrown out missing values (because we are controlling for missingness), but I can use the full data. ```r summary(lm(indegree~parentinc, data=addhealth))$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 3.93119831 0.106835499 36.796742 1.886044e-249 ## parentinc 0.01461554 0.001890363 7.731602 1.392430e-14 ``` ```r summary(lm(indegree~parentinc.meani+is.na(parentinc), data=addhealth))$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 3.93119831 0.105945249 37.105942 2.046037e-262 ## parentinc.meani 0.01461554 0.001874611 7.796570 7.888129e-15 ## is.na(parentinc)TRUE -0.18735131 0.130692401 -1.433529 1.517780e-01 ``` - This model still assumes MCAR and reduces variance in the independent variable. - Its primary advantage is that it is a quick method to avoid having to throw out cases that have valid data on other important variables. --- ## Mean imputation with dummy, Add Health <img src="module6_slides_model_complications_files/figure-html/missingdummy_plot-1.png" width="864" style="display: block; margin: auto;" /> --- ## Regression imputation - I will predict the value of parental income by other independent variables (but never the dependent variable) using a regression model. - In this case, I will transform parental income by the square root as well since it is heavily right skewed. ```r model <- lm(sqrt(parentinc)~race+pseudoGPA+honorsociety+alcoholuse+smoker +bandchoir+academicclub+nsports, data=addhealth) ``` -- Then, I use the `predict` command to get predicted values for all observations and impute the predicted values (or their square, technically) for missing values. ```r predicted <- predict(model, addhealth) addhealth$parentinc.regi <- addhealth$parentinc incmiss <- is.na(addhealth$parentinc) addhealth$parentinc.regi[incmiss] <- predicted[incmiss]^2 summary(addhealth$parentinc.regi) ``` ``` ## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's ## 0.00 25.19 39.83 43.87 54.20 200.00 39 ``` I still have some missing values, because there were missing values on the variables I used to predict parental income. --- ## Random regression imputation The previous model is deterministic, and will underestimate variance in parental income but I can add a random component to this by sampling from a normal distribution with a mean of zero and standard deviation equal to that of the model residuals. ```r addhealth$parentinc.rregi <- addhealth$parentinc addhealth$parentinc.rregi[incmiss] <- (predicted[incmiss]+ * rnorm(sum(incmiss), 0, sigma(model)))^2 sd(addhealth$parentinc, na.rm=TRUE) ``` ``` ## [1] 33.6985 ``` ```r sd(addhealth$parentinc.regi, na.rm=TRUE) ``` ``` ## [1] 30.10024 ``` ```r sd(addhealth$parentinc.rregi, na.rm=TRUE) ``` ``` ## [1] 32.80286 ``` --- ## Regression imputation, Add Health .pull-left[ <img src="module6_slides_model_complications_files/figure-html/regimpute_plot-1.png" width="504" style="display: block; margin: auto;" /> ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/rregimpute_plot-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Chained equations As you saw above, predictive imputation still leads to some missing values if the variables used for prediction also contain missing values. We can get around this via an iterative procedure called **chained equations**: -- 1. All missing values are given some placeholder value. This might be the mean value, for example. -- 2. For one variable, the placeholder values are removed and missing values put back in. These missing values are then predicted and imputed by some model for this variable. -- 3. Step 2 is repeated for all variables with missing values. When all variables have been imputed, we have completed one iteration. -- 4. Steps 2-3 are then repeated again for some number of iterations. The number of iterations necessary may vary by data, but five iterations is typical. --- ## Using `mice` to impute a single dataset The `mice` command will use chained equations to impute missing values on *all* variables when a dataset is fed in. I can then use the `complete` command to extract a full dataset with no missing values. ```r library(mice) imputed <- mice(addhealth[,c("indegree","race","sex","grade","pseudoGPA","honorsociety", "alcoholuse","smoker","bandchoir","academicclub","nsports", "parentinc")], m=1, print=FALSE) addhealth.ce <- complete(imputed, 1) apply(is.na(addhealth.ce), 2, sum) ``` ``` ## indegree race sex grade pseudoGPA honorsociety ## 0 0 0 0 0 0 ## alcoholuse smoker bandchoir academicclub nsports parentinc ## 0 0 0 0 0 0 ``` I can then use this new dataset `addhealth.ce` to run my models. --- ## Comparison of methods .stargazer[ <table cellspacing="0" align="center" style="border: none;"> <caption align="top" style="margin-bottom:0.3em;">Models predicting number of friend nominations, Add Health</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>deletion</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>mean</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>mean + dummy</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>random</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>regression</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>random regression</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>chained equations</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">3.391<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.360<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.399<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.470<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.350<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.441<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.340<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.120)</td> <td style="padding-right: 12px; border: none;">(0.111)</td> <td style="padding-right: 12px; border: none;">(0.115)</td> <td style="padding-right: 12px; border: none;">(0.104)</td> <td style="padding-right: 12px; border: none;">(0.107)</td> <td style="padding-right: 12px; border: none;">(0.105)</td> <td style="padding-right: 12px; border: none;">(0.103)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Parental income (1000s)</td> <td style="padding-right: 12px; border: none;">0.012<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.012<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.012<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.009<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.013<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.010<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.013<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> <td style="padding-right: 12px; border: none;">(0.002)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Smoker</td> <td style="padding-right: 12px; border: none;">0.383<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">0.201</td> <td style="padding-right: 12px; border: none;">0.203</td> <td style="padding-right: 12px; border: none;">0.196</td> <td style="padding-right: 12px; border: none;">0.187</td> <td style="padding-right: 12px; border: none;">0.193</td> <td style="padding-right: 12px; border: none;">0.201</td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.188)</td> <td style="padding-right: 12px; border: none;">(0.161)</td> <td style="padding-right: 12px; border: none;">(0.161)</td> <td style="padding-right: 12px; border: none;">(0.161)</td> <td style="padding-right: 12px; border: none;">(0.162)</td> <td style="padding-right: 12px; border: none;">(0.162)</td> <td style="padding-right: 12px; border: none;">(0.159)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Drinker</td> <td style="padding-right: 12px; border: none;">0.663<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.666<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.668<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.679<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.665<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.665<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.656<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.180)</td> <td style="padding-right: 12px; border: none;">(0.155)</td> <td style="padding-right: 12px; border: none;">(0.155)</td> <td style="padding-right: 12px; border: none;">(0.155)</td> <td style="padding-right: 12px; border: none;">(0.155)</td> <td style="padding-right: 12px; border: none;">(0.156)</td> <td style="padding-right: 12px; border: none;">(0.154)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Number of sports</td> <td style="padding-right: 12px; border: none;">0.454<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.476<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.474<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.481<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.462<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.475<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.461<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.049)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> <td style="padding-right: 12px; border: none;">(0.043)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Income missing</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-0.162</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.130)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.048</td> <td style="border-top: 1px solid black;">0.046</td> <td style="border-top: 1px solid black;">0.046</td> <td style="border-top: 1px solid black;">0.044</td> <td style="border-top: 1px solid black;">0.048</td> <td style="border-top: 1px solid black;">0.045</td> <td style="border-top: 1px solid black;">0.049</td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">3332</td> <td style="border-bottom: 2px solid black;">4343</td> <td style="border-bottom: 2px solid black;">4343</td> <td style="border-bottom: 2px solid black;">4343</td> <td style="border-bottom: 2px solid black;">4320</td> <td style="border-bottom: 2px solid black;">4320</td> <td style="border-bottom: 2px solid black;">4397</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="9"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] --- ## Imputation variability -- - The methods of random imputation have the benefit of preserving the standard deviation of the imputed variable and therefore calculating correct standard errors, but they also introduce a new source of uncertainty. -- - Each time I do an imputation with a random component (e.g. random regression, chained equation), I will get a somewhat different set of values. -- - Therefore, we now have **imputation variability** to add to our inferential concerns alongside **sampling variability**. -- - We can use **multiple imputation** to adjust our results for **imputation variability**. --- ## Multiple imputation process -- 1. Use imputation process with random component to impute missing values and repeat this process to produce `\(m\)` separate **complete datasets**. Each of these datasets will be somewhat different due to the randomization of imputation. Usually `\(m=5\)` is sufficient. -- 2. Run `\(m\)` separate parallel models on each imputed **complete dataset**. As a result, you will have `\(m\)` sets of regression coefficients and standard errors. -- 3. Pool the regression coefficients across datasets by taking the mean across all `\(m\)` datasets. -- 4. Pool standard errors by taking the mean across all `\(m\)` datasets *plus* the between model standard deviation in coefficients. The formula for the overall standard error is: `$$SE_{\beta}=\sqrt{W+(B+\frac{B}{m})}$$` Where `\(W\)` is the squared mean standard error across all `\(m\)` datasets, and `\(B\)` is the variance in coefficient estimates calculated across all `\(m\)` models. --- ## Multiple imputation with the `mice` package To get multiple complete datasets with `mice` we just need to specify the number of distinct complete datasets to create in the second argument: ```r imputations <- mice(popularity, 5, printFlag=FALSE) ``` -- - The imputations object now contains five fully complete imputed datasets. Its possible to extract any one of these datasets with the command `complete(imputations, i)` where `i` is replaced by a number between 1 and 5. -- - We can now conduct our parallel analysis on these five datasets and combine results. There is easy and a hard way to do this. Its useful to know both. --- ## Easy way: let `mice` do the hard work The `mice` package has a lot of nice features, including an object specific function for the `with` command and a `pool` command that make multiple imputation as easy as falling off a log: ```r model.mi <- pool(with(imputations, lm(indegree~parentinc+smoker+alcoholuse+nsports))) summary(model.mi) ``` ``` ## estimate std.error statistic df p.value ## (Intercept) 3.38654549 0.102909528 32.907988 4389.514 0.000000e+00 ## parentinc 0.01126339 0.001630526 6.907827 4389.514 5.627498e-12 ## smokerSmoker 0.23538273 0.159687599 1.474020 4389.514 1.405479e-01 ## alcoholuseDrinker 0.64050221 0.154054327 4.157639 4389.514 3.276895e-05 ## nsports 0.47096906 0.042956107 10.963961 4389.514 0.000000e+00 ``` --- ## Hard way: for-loop The hard way isn't really that hard. It is useful to know for cases where the easy way won't work, such as when you need to run complicated models (using `svydesign` would be an example). ```r b <- se <- NULL for(i in 1:5) { imputation <- complete(imputations,i) model <- lm(indegree~parentinc+smoker+alcoholuse+nsports, data=imputation) b <- cbind(b, coef(model)) se <- cbind(se, summary(model)$coef[,2]) } b ``` ``` ## [,1] [,2] [,3] [,4] [,5] ## (Intercept) 3.38654549 3.38654549 3.38654549 3.38654549 3.38654549 ## parentinc 0.01126339 0.01126339 0.01126339 0.01126339 0.01126339 ## smokerSmoker 0.23538273 0.23538273 0.23538273 0.23538273 0.23538273 ## alcoholuseDrinker 0.64050221 0.64050221 0.64050221 0.64050221 0.64050221 ## nsports 0.47096906 0.47096906 0.47096906 0.47096906 0.47096906 ``` The `b` and `se` objects are matrices that contain the coefficients and standard errors, respectively, for each model on the column. --- ## Hard way: pool the results Now we can pool the results using the `b` and `se` matrices and some creative use of `apply` commands. ```r b.pool <- apply(b,1,mean) between.var <- apply(b,1,var) within.var <- apply(se^2,1,mean) se.pool <- sqrt(within.var+between.var+between.var/5) t.pool <- b.pool/se.pool pvalue.pool <- (1-pnorm(abs(t.pool)))*2 data.frame(b.pool, se.pool, t.pool, pvalue.pool) ``` ``` ## b.pool se.pool t.pool pvalue.pool ## (Intercept) 3.38654549 0.102909528 32.907988 0.000000e+00 ## parentinc 0.01126339 0.001630526 6.907827 4.921397e-12 ## smokerSmoker 0.23538273 0.159687599 1.474020 1.404762e-01 ## alcoholuseDrinker 0.64050221 0.154054327 4.157639 3.215542e-05 ## nsports 0.47096906 0.042956107 10.963961 0.000000e+00 ``` --- class: inverse, center, middle background-image: url(images/nareeta-martin-z7LRp54Yulw-unsplash.jpg) background-size: cover name: multicollinearity # Multicollinearity and Scale Creation --- ## The problem of multicollinearity - Given that adding more independent variables to your model allows you to account for potential omitted variable bias, why wouldn't you just put in as many variables as you can? -- - Because of **multicollinearity**. Multicollinearity occurs when there is collectively a high correlation between the independent variables in the model. -- - Intuitively, its easy to understand why multicollinearity is a problem. When two independent variables are highly correlated with one another, it becomes hard to separate out their effect on the dependent variable. -- - Technically, the effect of multicollinearity is to inflate standard errors and make coefficient estimates highly unstable across different combinations of highly collinear terms in the model. --- ## Two kinds of multicollinearity -- .pull-left[ ### Structural multicollinearity Structural multicollinearity occurs when one independent variable is completely determined by another independent variable or set of independent variables. This really isn't an issue with the data, but rather a specification error by the researcher. ] -- .pull-right[ ### Data-based multicollinearity Data-based multicollinearity occurs when a set of variables are highly but not perfectly correlated with one another in the empirical data. It is a hazard of using observational data where the characteristics for which you want to get separate effects are difficult to separate. ] --- ## Example of structural multicollinearity Lets predict violent crime by % female and % male. ```r crimes$PctFemale <- 100-crimes$PctMale summary(lm(Violent~PctMale+PctFemale, data=crimes)) ``` ``` ## ## Call: ## lm(formula = Violent ~ PctMale + PctFemale, data = crimes) ## ## Residuals: ## Min 1Q Median 3Q Max ## -293.57 -104.01 -54.44 88.98 783.97 ## *## Coefficients: (1 not defined because of singularities) ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4317.26 1628.20 2.652 0.0108 * ## PctMale -79.75 33.01 -2.416 0.0195 * *## PctFemale NA NA NA NA ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 186.5 on 49 degrees of freedom ## Multiple R-squared: 0.1064, Adjusted R-squared: 0.08819 ## F-statistic: 5.836 on 1 and 49 DF, p-value: 0.01948 ``` --- ## Singularity! Not as cool as it sounds. One of the terms was dropped from the model because the terms are perfectly collinear. ```r cor(crimes$PctFemale,crimes$PctMale) ``` ``` ## [1] -1 ``` -- This is not a problem of the model, but our thinking. Either term by itself will give you full information. ```r coef(lm(Violent~PctMale, data=crimes)) ``` ``` ## (Intercept) PctMale ## 4317.26466 -79.75285 ``` ```r coef(lm(Violent~PctFemale, data=crimes)) ``` ``` ## (Intercept) PctFemale ## -3658.02067 79.75285 ``` --- ## Detecting data-based multicollinearity .left-column[  ] .right-column[ - Standard errors increase substantially across nested models with more covariates - Regression coefficients are highly unstable across nested models - Examination of correlation matrix - Calculate variance inflation factors ] --- ## Multicollinearity example: NYC non-profits .pull-left[ We will look at data collected by myself and Nicole Marwell on the spatial distribution of money contracted out to organization for social services by the City of New York from 1997-2001. The unit of analysis is a NYC health area, which can loosely be thought of as a neighborhood. The variables are: - **amountcapita**: The dollar amount of money provided to the health area divided by the population size of the health area. We will log it due to skewness. - **poverty**: percent of population below the poverty line. - **unemployed**: unemployment rate. - **income**: median household income. We will log it due to skewness. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/nyc-map-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Model results, NYC example .right-column[ .stargazer[ <table cellspacing="0" align="center" style="border: none;"> <caption align="top" style="margin-bottom:0.3em;">Models predicting social service funding per capita (logged)</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 1</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 2</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 3</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 4</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 5</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 6</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 7</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">intercept</td> <td style="padding-right: 12px; border: none;">4.14<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.13<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">10.24<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">4.12<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">-24.70<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">3.08</td> <td style="padding-right: 12px; border: none;">-29.60<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.17)</td> <td style="padding-right: 12px; border: none;">(0.23)</td> <td style="padding-right: 12px; border: none;">(2.05)</td> <td style="padding-right: 12px; border: none;">(0.23)</td> <td style="padding-right: 12px; border: none;">(4.94)</td> <td style="padding-right: 12px; border: none;">(3.27)</td> <td style="padding-right: 12px; border: none;">(5.31)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">poverty rate</td> <td style="padding-right: 12px; border: none;">0.04<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.04<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.13<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.12<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.01)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.01)</td> <td style="padding-right: 12px; border: none;">(0.02)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.02)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">unemployment rate</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.09<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.00</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">0.10<sup style="vertical-align: 0px;">**</sup></td> <td style="padding-right: 12px; border: none;">0.08<sup style="vertical-align: 0px;">*</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.02)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.03)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.04)</td> <td style="padding-right: 12px; border: none;">(0.03)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">median income (logged)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-0.49<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">2.52<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">0.09</td> <td style="padding-right: 12px; border: none;">2.91<sup style="vertical-align: 0px;">***</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.19)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(0.43)</td> <td style="padding-right: 12px; border: none;">(0.28)</td> <td style="padding-right: 12px; border: none;">(0.46)</td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.08</td> <td style="border-top: 1px solid black;">0.04</td> <td style="border-top: 1px solid black;">0.02</td> <td style="border-top: 1px solid black;">0.08</td> <td style="border-top: 1px solid black;">0.16</td> <td style="border-top: 1px solid black;">0.04</td> <td style="border-top: 1px solid black;">0.18</td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> <td style="border-bottom: 2px solid black;">341</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="9"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] ] .left-column[ The models exhibit classic signs of multicollinearity: - Highly unstable coefficients across models. - Large increase in standard errors. ] --- ## The correlation matrix as diagnostic .pull-left[ ```r cor(nyc[,c("poverty","unemployed","lincome")]) ``` ``` ## poverty unemployed lincome ## poverty 1.0000000 0.6982579 -0.9120680 ## unemployed 0.6982579 1.0000000 -0.7419472 ## lincome -0.9120680 -0.7419472 1.0000000 ``` The correlation between the three variables is very high, suggesting strong multicollinearity. Note that, while the correlation matrix is often helpful, it may not reveal the full extent of multicollinearity because it only looks at bivariate relationships between the variables. ] -- .pull-right[ ### Visualize as correlogram ```r library(corrgram) corrgram(nyc[,c("poverty","unemployed", "lincome")], upper.panel="panel.cor", lower.panel="panel.pts") ``` <img src="module6_slides_model_complications_files/figure-html/corrgram-nyc-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Variance inflation factors The **variance inflation factor** (VIF) is the multiplicative factor by which the variance in the estimation of any one coefficient from the current model is increased due to multicollinearity relative to a model in which the variables are uncorrelated. The square root of the VIF is roughly the expected factor increase in the standard error. It can be shown that the VIF for the `\(i\)`th variable is given by: `$$VIF_i=\frac{1}{1-R_i^2}$$` Where `\(R_i^2\)` is the r-squared value when the given independent variable is predicted by all of the other independent variables in the model. For example, we could calculate the VIF for poverty in the full model by: ```r 1/(1-summary(lm(poverty~unemployed+lincome, data=nyc))$r.squared) ``` ``` ## [1] 5.984485 ``` The square root of this VIF is 2.45, indicating that the standard error for poverty is more than doubled due to multicollinearity in the full model. --- ## Estimating VIF with the `vif` command We can also use the `vif` function in the `car` package to quickly estimate all VIFs for a given model: ```r library(car) vif(model.allthree) ``` ``` ## poverty unemployed lincome ## 5.984485 2.238379 6.822173 ``` The general rule of thumb is that a VIF over four is problematic enough that it needs to be addressed in some manner. Clearly, multicollinearity is a big problem here. --- ## You have multicollinearity, now what? -- ### Remove variables A simple approach is to remove some of the highly correlated covariates. However, because the variables are not perfectly correlated, you are basically throwing out some information. -- ### Separate models Another approach is to run separate models with only one of the highly collinear variables in each model. This can also be unsatisfying because each of the models is underestimating the total effect of the variables collectively. -- ### Create a scale In cases where the collinear variables are all thought to represent the same underlying conceptual variable, we can combine them into a single synthetic scale. --- ## Standardization and reverse ordering .pull-left[ - In many cases, the variables that are thought to make up a scale might be measured in the same manner. - When variables are measured differently (as in our example), then they must be standardized in some way to make them comparable before scale construction and evaluation. The most common way to do this is by creating **z-scores** by subtracting by the mean and dividing by the standard deviation of each variable. - Some variables may be positively related to the underlying concept while others may be negatively related. It may be necessary to reverse the coding of a variable to make all variables positively related to the underlying concept. ] .pull-right[ ### NYC example ```r nyc$poverty.z<-(nyc$poverty-mean(nyc$poverty))/ sd(nyc$poverty) nyc$unemployed.z<-(nyc$unemployed-mean(nyc$unemployed))/ sd(nyc$unemployed) nyc$lincome.z<- -1*(nyc$lincome-mean(nyc$lincome))/ sd(nyc$lincome) ``` Note that I am multiplying the `lincome` result by -1 to reverse code it. I can also use the `scale` command to do this: ```r nyc$poverty.z <- scale(nyc$poverty) nyc$unemployed.z <- scale(nyc$unemployed) nyc$lincome.z <- -1*scale(nyc$lincome) ``` ] --- ## Cronbach's alpha Cronbach's `\(\alpha\)` is a statistic developed in psychology to test the degree to which different variables measure the same underlying concept. -- - Cronbach's `\(\alpha\)` is often thought of as a test of the **internal reliability** of a scale. -- - You can think about Cronbach's `\(\alpha\)` as a summary measure of the correlation matrix we saw earlier. It goes from 0 to 1, with 0 indicating no shared correlation, and 1 indicating perfect correlation between all items. -- The `pysch` package includes and `alpha` command that will calculate `\(\alpha\)`. ```r psych::alpha(nyc[,c("poverty.z","unemployed.z","lincome.z")])$total ``` ``` ## raw_alpha std.alpha G6(smc) average_r S/N ase mean ## 0.9159293 0.9159293 0.9013009 0.784091 10.89474 0.008313234 -2.211598e-16 ## sd median_r ## 0.9252355 0.7419472 ``` The `\(\alpha\)` of 0.915 indicates a very high level of shared covariance between the three variables. --- ## Summated scale Once I have standardized and (if necessary) reverse coded all of the variables for my scale, I can create a simple **summated scale** by adding them up. I am also going to scale this variable with the `scale` command so that it has a mean of zero and a standard deviation of one. ```r nyc$deprivation_summated <- scale(nyc$poverty.z+nyc$unemployed.z+nyc$lincome.z) model <- lm(log(amtcapita)~deprivation_summated, data=nyc) summary(model)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.9474224 0.08807143 56.175114 8.329185e-174 ## deprivation_summated 0.3747882 0.08820085 4.249258 2.775153e-05 ``` ```r exp(coef(model)) ``` ``` ## (Intercept) deprivation_summated ## 140.811538 1.454683 ``` A one standard deviation increase on my deprivation scale is associated with a 45% increase in the amount for social services in a neighborhood. --- ## Factor analysis Another approach to creating a scale is **factor analysis**. Factor analysis is a method to extract the underlying latent variables (the factors) from a set of observed variables. We will start with an example where we assume a single underlying latent factor. -- Lets say we have three variables `\(z_1\)`, `\(z_2\)`, and `\(z_3\)` all measured as z-scores. We can construct a system of equations that relate each of these variables to a common shared factor `\(F\)` (also on a standard scale with a mean of 0 and SD of 1) and three unique factors, `\(Q_1\)`, `\(Q_2\)`, and `\(Q_3\)`. `$$z_{i1}=b_1F_i+u_1Q_{i1}$$` `$$z_{i2}=b_2F_i+u_2Q_{i2}$$` `$$z_{i3}=b_3F_i+u_3Q_{i3}$$` The `\(F_i\)` are called the **factor scores** and are the values of the common factor for each observation. The `\(b\)` values are called the **factor loadings** and give the correlation between each observed variable and the common factor. The unique factor components are not actually estimated in factor analysis but serve as the "residual" components. --- ## Estimation in factor analysis .pull-left[ The factor scores and factor loadings for a factor analysis can be estimated by mathematical manipulation of the observed correlation matrix between variables, but we won't get into the mathematical details here. - The `factanal` command in base R will estimate a factor analysis by maximum likelihood estimation (a technique we will learn in the next module). - The `fa` command in the `psych` package will also estimate factor analysis using a variety of techniques. It also has some nice additional tools that will make it more useful. ] -- .pull-right[ ### NYC example ```r factor_nyc <- fa(nyc[,c("poverty", "unemployed", "lincome")], 1, rotate="oblimin") ``` - The second argument specifies that we want only one common factor. - We also need to specify a technique of *rotation* for the factor loadings because there are an infinite number of possible ways to express them. The oblimin method is a standard approach that helps to maximize differences between factors without forcing them to be uncorrelated. ] --- ## Factor loadings .pull-left[ ### Extract loadings ```r loadings(factor_nyc) ``` ``` ## ## Loadings: ## MR1 ## poverty 0.926 ## unemployed 0.754 ## lincome -0.984 ## ## MR1 ## SS loadings 2.396 ## Proportion Var 0.799 ``` The bottom part shows that 79.9% of the variation in the three variables is accounted for by the common factor. The loadings themselves show the correlation coefficient between each observed variable and the common factor. ] -- .pull-right[ ### Visualize factor loadings ```r fa.diagram(factor_nyc) ``` <img src="module6_slides_model_complications_files/figure-html/visualize-factor-loading-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Factor Scores I can easily extract my factor scores from the the factor analysis object and use them as my scale measure of deprivation. ```r nyc$deprivation_factor <- factor_nyc$scores[,"MR1"] model <- lm(log(amtcapita)~deprivation_factor, data=nyc) summary(model)$coef ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 4.9474224 0.08908910 55.533419 2.797390e-172 ## deprivation_factor 0.2849722 0.09036319 3.153631 1.756654e-03 ``` ```r exp(coef(model)) ``` ``` ## (Intercept) deprivation_factor ## 140.811538 1.329725 ``` A one standard deviation increase on my deprivation scale is associated with a 33% increase in the amount for social services in a neighborhood. --- ## Whats the difference? .pull-left[ <img src="module6_slides_model_complications_files/figure-html/compare-factor-summated-1.png" width="504" style="display: block; margin: auto;" /> ] -- .pull-right[ - The summated scale uses all the variation in the three variables to generate the score. - The factor analysis only uses the shared component to generate the score. The unique component is left out. - A closely related technique to factor analysis called **principal component analysis** uses all of the variation in the items and will produce results virtually identical to the summated score. ] --- ## Factor analysis with multiple factors The previous example only used one factor and produced results that were very similar to a simple summated scale. However, it is also possible to use factor analysis to identify more than one common factor shared among a set of variables. -- The formulas for factor analysis above can be generalized to a set of `\(J\)` observed variables and `\(m\)` factors by a set of `\(J\)` equations. For the `\(j\)`th observed variable: `$$z_{ji}=b_{j1}F_{1i}+b_{j2}F_{2i}+\ldots+b_{jm}F_{ji}+u_jQ_{ji}$$` There will be a set of `\(J\)` factor loadings for each of the `\(m\)` factors. The key question with this technique is what are the appropriate number of factors? This is typically determined by an analysis of: -- - how much total variation is explained by a given number of factors. -- - whether the factor loadings for a given number of factors make theoretical sense. --- ## Example: social conservatism among Muslims .pull-left[ Between 2008-12, The Pew Research Center surveyed Muslims from numerous countries around the world and (in addition to other questions) asked respondents about the moral acceptability of these practices: - divorce - polygamy - fertility control - alcohol - euthanasia - suicide - abortion - prostitution - premarital sex - homosexuality ] -- .pull-right[ The data are coded as an ordinal response: - Morally acceptable - Depends/Not a moral issue - Morally wrong I re-code the data on a numeric scale from 1 to 3. ```r morality <- cbind(divorce=pew$moral_divorce, fertility=pew$moral_fertility, alcohol=pew$moral_alcohol, euthansia=pew$moral_euthansia, suicide=pew$moral_suicide, abortion=pew$moral_abortion, prostitution=pew$moral_prostitution, premar_sex=pew$moral_premar_sex, moral_gay=pew$moral_gay) ``` ] --- ## Correlogram of responses ```r corrgram(morality, upper.panel="panel.cor", order="PCA") ``` <img src="module6_slides_model_complications_files/figure-html/correlogram-moral-1.png" width="864" style="display: block; margin: auto;" /> --- ## Using the correlation matrix as data -- - Factor analysis only needs the correlation matrix between variables rather than the original data itself. -- - This is one case where available-case analysis makes sense. I can maximize the number of cases on each pairwise correlation coefficient to use all of the available data. -- The `use="pairwise.complete.obs"` argument in the `cor` function will use available-case analysis when it computes the correlation coefficient between each pair of variables. ```r morality_r <- cor(morality, use="pairwise.complete.obs") ``` -- ### Try factor analysis with up to three factors ```r morality_fa1 <- fa(morality_r, 1, rotate="oblimin") morality_fa2 <- fa(morality_r, 2, rotate="oblimin") morality_fa3 <- fa(morality_r, 3, rotate="oblimin") ``` --- ## Comparing models with `fa.diagram` <img src="module6_slides_model_complications_files/figure-html/nfactors-1.png" width="864" style="display: block; margin: auto;" /> --- ## Extending factor analysis -- Factor analysis is part of a larger family of methods that are often described as **data reduction** techniques: How can you reduce the amount of information in a given set of variables into a smaller set of variables? -- .pull-left[ ### Latent class analysis An important cousin of factor analysis that can be used for categorical variables. ] -- .pull-right[ ### Structural equation modeling The idea of factor analysis can be generalized to construct entire systems of equations that involve both latent and observed variables. ] --- class: inverse, center, middle background-image: url(images/caleb-jones-J3JMyXWQHXU-unsplash.jpg) background-size: cover name: model-selection # Model Selection --- ## Predicting violent crime rates Lets predict violent crime rates in US States by a variety of characteristics. Note that some of these models are **nested** within other models, meaning that the more complex models just add additional variables to earlier models. Other models are non-nested. ```r model.demog <- lm(Violent~MedianAge+PctMale, data=crimes) model.ineq <- update(model.demog,.~.+Gini) model.pov <- update(model.ineq,.~.+Poverty) model.unemp <- update(model.pov,.~.+Unemployment) model.justunemp <- lm(Violent~Unemployment, data=crimes) model.another <- update(model.justunemp,.~.+Gini+MedianAge) ``` --- ## Which model is best? .stargazer[ <table cellspacing="0" align="center" style="border: none;"> <caption align="top" style="margin-bottom:0.3em;">Models predicting violent crime rate</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 1</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 2</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 3</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 4</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 5</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 6</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">7534.81<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">619.07</td> <td style="padding-right: 12px; border: none;">604.71</td> <td style="padding-right: 12px; border: none;">128.07</td> <td style="padding-right: 12px; border: none;">-6.09</td> <td style="padding-right: 12px; border: none;">-638.35</td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(1724.03)</td> <td style="padding-right: 12px; border: none;">(2995.68)</td> <td style="padding-right: 12px; border: none;">(3031.21)</td> <td style="padding-right: 12px; border: none;">(2972.28)</td> <td style="padding-right: 12px; border: none;">(113.33)</td> <td style="padding-right: 12px; border: none;">(599.66)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Median Age</td> <td style="padding-right: 12px; border: none;">-38.56<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">-27.84<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">-28.06<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">-27.84<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-24.90<sup style="vertical-align: 0px;">*</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(10.89)</td> <td style="padding-right: 12px; border: none;">(10.94)</td> <td style="padding-right: 12px; border: none;">(11.28)</td> <td style="padding-right: 12px; border: none;">(11.02)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(9.40)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Percent Male</td> <td style="padding-right: 12px; border: none;">-115.67<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">-23.28</td> <td style="padding-right: 12px; border: none;">-23.21</td> <td style="padding-right: 12px; border: none;">-12.29</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(31.39)</td> <td style="padding-right: 12px; border: none;">(44.75)</td> <td style="padding-right: 12px; border: none;">(45.23)</td> <td style="padding-right: 12px; border: none;">(44.59)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;">Gini</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">43.01<sup style="vertical-align: 0px;">**</sup></td> <td style="padding-right: 12px; border: none;">43.71<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">39.04<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">38.68<sup style="vertical-align: 0px;">**</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(15.68)</td> <td style="padding-right: 12px; border: none;">(17.36)</td> <td style="padding-right: 12px; border: none;">(17.15)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(11.67)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Poverty Rate</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-0.88</td> <td style="padding-right: 12px; border: none;">-6.09</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(8.91)</td> <td style="padding-right: 12px; border: none;">(9.17)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;">Unemployment Rate</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">25.47</td> <td style="padding-right: 12px; border: none;">45.89<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">23.02</td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(14.18)</td> <td style="padding-right: 12px; border: none;">(12.99)</td> <td style="padding-right: 12px; border: none;">(13.14)</td> </tr> <tr> <td style="border-top: 1px solid black;">R<sup style="vertical-align: 0px;">2</sup></td> <td style="border-top: 1px solid black;">0.29</td> <td style="border-top: 1px solid black;">0.39</td> <td style="border-top: 1px solid black;">0.39</td> <td style="border-top: 1px solid black;">0.43</td> <td style="border-top: 1px solid black;">0.20</td> <td style="border-top: 1px solid black;">0.42</td> </tr> <tr> <td style="border-bottom: 2px solid black;">Num. obs.</td> <td style="border-bottom: 2px solid black;">51</td> <td style="border-bottom: 2px solid black;">51</td> <td style="border-bottom: 2px solid black;">51</td> <td style="border-bottom: 2px solid black;">51</td> <td style="border-bottom: 2px solid black;">51</td> <td style="border-bottom: 2px solid black;">51</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="8"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] --- ## A tradeoff: accuracy vs. parsimony .pull-left[ .center[] ### Accuracy We want our models to be accurate, or more formally we want them to have high **goodness of fit**. This is measured in OLS regression models primarily by the `\(R^2\)` value. A more accurate model accounts for more of the variation in the dependent variable. ] -- .pull-right[ .center[] ### Parsimony We want our models to be parsimonious. They should account for variation in the dependent variable with a minimal number of explanatory variables. Models that are not parsimonious are rarely theoretically very interesting. ] ??? We can always add more variables, interactions, non-linear terms, and so forth to get a more accurate model, but this comes at the cost of parsimony. Ultimately, this would lead to us simply throwing up our hands and saying "everybody is different." We knew that already. Our goal is to see if a substantial part of the observed variation in outcome can be explained by a minimal set of predictors, in theoretically expected (or sometimes unexpected) ways. --- ## How to choose a model -- .pull-left[ ### Do Nots - **Do not** choose a model based on all of the coefficients being statistically significant. That is a misuse of statistical inference based on an incorrect interpretation of a p-value. - **Do not** choose a model because it produces the results you like the best. That is not science. - **Do not** choose a model based on some purely mechanical algorithm. Model choice needs to be driven by what questions you want to ask as much as goodness of fit and parsimony. - **Do not** choose a model based on `\(R^2\)` values. An additional variable will make the `\(R^2\)` value somewhat better, so there is no end to the complexity of the model you fit. ] -- .pull-right[ ### Good Practices - **Do** think theoretically about what is the best model for the question at hand. - **Do** make use of model selection tools to help make a decision, but not in a mechanistic way. Use multiple tools. - **Do** conduct and report sensitivity analyses showing how dependent your result is upon the selected model vs. other models. - **Do** weigh both parsimony and accuracy, relative to the goals of your research question. ] ??? Maybe the control variable needs to be in there because prior work or theory says its important, regardless of what the model fit statistics say. Perhaps interactions need to be in the model because your key interest is in intersectionality. And so on. --- ## Model selection tools - The F-Test - Adjusted `\(R^2\)` - Bayesian Information Criterion (BIC) - Bayesian Model Averaging (BMA) --- ## Null vs. saturated model Many model selection tools utilize a distinction between the **null model** and the **saturated model**. -- .pull-left[ ### Null model `$$\hat{y}_i=\bar{y}$$` The null model is the regression model with no predictors. This model is the most parsimonious, but the least accurate. ] -- .pull-right[ ### Saturated model `$$\hat{y}_i=\beta_0+\beta_1x_{i1}+\beta_2x_{i2}+\ldots+\beta_px_{ip}$$` Where, `$$n=p$$` The saturated model is a hypothetical model in which the number of predictors equals the number of observations and we fit each `\(y_i\)` exactly. This model is the most accurate with `\(R^2=1\)`, but the least parsimonious. ] --- ## The F-Test The F-test is a classical hypothesis test for comparing two different models in which one model is nested within the other. This means that the second model includes all of the same independent variables as model 1 as well as an additional `\(g\)` independent variables. The null hypothesis for the F-test is that the effects for all of the additional `\(g\)` variables are zero. -- The F-test produces a test statistic called the **F-statistic**: `$$F=\frac{(SSR_1-SSR_2)/g}{SSR_2/(n-g-k-1)}$$` Where `\(SSR\)` is the sum of squared residuals for a given model, `\(g\)` is the additional terms added to the second model, and `\(k\)` is the number of terms in the first model. The F-statistic is a ratio of the mean amount of variation explained by the new predictors in the second model (numerator) by the mean amount of variation unexplained per degree of freedom in the second model (denominator). -- Assuming the null hypothesis is true, the F-statistic will come from an F-distribution with degrees of freedom equal to `\(g\)` and `\(n-g-k-1\)` (the F-distribution has two DF parameters). Thus, its possible to determine how far out in the tail of this distribution the estimated F-statistic is to calculate a p-value for the hypothesis test. --- ## Calculating F-statistic .pull-left[ ```r model.null <- lm(Violent~1, data=crimes) SSR1 <- sum(model.null$residuals^2) SSR2 <- sum(model.demog$residuals^2) g <- length(model.demog$coef)-length(model.null$coef) k <- length(model.null$coef)-1 n <- length(model.null$residuals) Fstat <- ((SSR1-SSR2)/g)/(SSR2/(n-g-k-1)) Fstat ``` ``` ## [1] 9.869375 ``` ```r 1-pf(Fstat, g, n-g-k-1) ``` ``` ## [1] 0.0002568606 ``` The p-value for the hypothesis test is 0.00026. So, I would likely not reject the null hypothesis and would prefer the null model to the model predicting violent crime by median age and percent male. ] .pull-right[ <img src="module6_slides_model_complications_files/figure-html/anovaplot-1.png" width="504" style="display: block; margin: auto;" /> ] --- ## Using `anova` command for F-test Rather than do this by hand, you can feed both models into the `anova` command. ```r anova(model.null, model.demog) ``` ``` ## Analysis of Variance Table ## ## Model 1: Violent ~ 1 ## Model 2: Violent ~ MedianAge + PctMale ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 50 1907432 ## 2 48 1351616 2 555817 9.8694 0.0002569 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- ## Multiple model comparisons The `anova` command is not limited to two models. We can feed in a list of models **so long as they are nested models.** ```r anova(model.null, model.demog, model.ineq, model.unemp) ``` ``` ## Analysis of Variance Table ## ## Model 1: Violent ~ 1 ## Model 2: Violent ~ MedianAge + PctMale ## Model 3: Violent ~ MedianAge + PctMale + Gini ## Model 4: Violent ~ MedianAge + PctMale + Gini + Poverty + Unemployment ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 50 1907432 ## 2 48 1351616 2 555817 11.5056 9.209e-05 *** ## 3 47 1165120 1 186496 7.7211 0.007935 ** ## 4 45 1086941 2 78179 1.6183 0.209554 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` The F-statistic is somewhat different than the two model comparison because the denominator for each F-statistic always uses the most complex model. --- ## F-test for adding one variable ```r anova(model.demog, model.ineq) ``` ``` ## Analysis of Variance Table ## ## Model 1: Violent ~ MedianAge + PctMale ## Model 2: Violent ~ MedianAge + PctMale + Gini ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 48 1351616 ## 2 47 1165120 1 186496 7.5231 0.008593 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ```r summary(model.ineq)$coef["Gini",] ``` ``` ## Estimate Std. Error t value Pr(>|t|) ## 43.006211344 15.679510943 2.742828619 0.008593105 ``` The p-value for an F-test where only one variable is added is identical to the p-value for a t-statistic of that particular regression coefficient. So, the F-test really only adds something when comparing across models where the more complex model adds multiple new variables. --- ## Limitations of the F-test -- - The F-test can only be used to compare nested models. This limits its use substantially because we can imagine that it might be theoretically important to compare non-nested models. -- - The F-test for adding a single variable is equal to the same t-test for the regression coefficient of that variable in the new model. If we just add each new variable sequentially to our nested models, the F-test adds nothing new. If we add in clusters of variables, the F-test does not help us distinguish which of the variables in the cluster were most important. -- - The F-test is based on the logic of null hypothesis significance testing. That means it is largely driven by sample size. The preferred model by the F-test will differ across two different datasets if they vary in sample size but are the same otherwise. --- ## Methods for penalizing goodness of fit -- - `\(R^2\)` can be thought of as a goodness-of-fit statistic. It measures the accuracy of the model in predicting the dependent variable. -- - `\(R^2\)` cannot be used directly to select models because it can only get bigger as more independent variables are added to the model. It will always prefer the more complex model unless there is literally zero correlation between the new independent variables and the dependent variable. -- - What if we could penalize `\(R^2\)` by some value that depends on the number of independent variables in the model? This would give us a single measure that combines accuracy and parsimony. --- ## Adjusted `\(R^2\)` The formula for adjusted `\(R^2\)` is: `$$R^2_{adj}=R^2-(\frac{p}{n-p-1})(1-R^2)$$` Where `\(p\)` is the number of independent variables in the model. The subtracted part is the parsimony penalty and it is a function of the number of variables as a proportion of the sample size and the lack of goodness-of-fit, `\(1-R^2\)`. -- This value is calculated by default by the `summary` command for a linear model in R. It also shows up in the default output to `texreg`. ```r summary(model.demog)$r.squared ``` ``` ## [1] 0.2913952 ``` ```r summary(model.demog)$adj.r.squared ``` ``` ## [1] 0.26187 ``` --- ## Bayesian Information Criterion There are two common forms of BIC: - BIC is implicitly compared to the saturated model. The `BIC` command in R will give you this value. - BIC' is implicitly compared to the null model. -- The equation for BIC' for OLS regression models is most intuitive: `$$BIC'=n\log(1-R^2)+p\log(n)$$` Where `\(p\)` is the number of independent variables in the model. The first component is the accuracy of the model and the second component is the parsimony penalty. -- BIC is unusual in that lower values are better, and negative values are most preferred. A negative value means you prefer this model to its comparison (e.g. null or saturated model). You can directly compare the values of any two given models by BIC' or BIC, and you will prefer the model with the lower value. The models do not need to be nested. --- ## Calculating BIC ```r bic.null <- function(model) { rsq <- summary(model)$r.squared n <- length(model$resid) p <- length(model$coef)-1 return(n*log(1-rsq)+p*log(n)) } bic.null(model.demog) ``` ``` ## [1] -9.703675 ``` ```r BIC(model.demog) ``` ``` ## [1] 679.8933 ``` ```r BIC(model.demog)-BIC(model.null) ``` ``` ## [1] -9.703675 ``` --- ## BIC or BIC' give the same difference ```r bic.null(model.unemp)-bic.null(model.demog) ``` ``` ## [1] 0.6808653 ``` ```r BIC(model.unemp)-BIC(model.demog) ``` ``` ## [1] 0.6808653 ``` -- We slightly prefer the more complex model with unemployment, poverty, and gini in addition to the demographic characteristics. -- A general rule of thumb is that BIC differences less than two give weak preferences, while BIC differences greater than six give strong preferences, and BIC differences greater than ten give very strong preferences. --- ## Model comparison .stargazer[ <table cellspacing="0" align="center" style="border: none;"> <caption align="top" style="margin-bottom:0.3em;">Models predicting violent crime rate</caption> <tr> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b></b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 1</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 2</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 3</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 4</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 5</b></th> <th style="text-align: left; border-top: 2px solid black; border-bottom: 1px solid black; padding-right: 12px;"><b>Model 6</b></th> </tr> <tr> <td style="padding-right: 12px; border: none;">Intercept</td> <td style="padding-right: 12px; border: none;">7534.81<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">619.07</td> <td style="padding-right: 12px; border: none;">604.71</td> <td style="padding-right: 12px; border: none;">128.07</td> <td style="padding-right: 12px; border: none;">-6.09</td> <td style="padding-right: 12px; border: none;">-638.35</td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(1724.03)</td> <td style="padding-right: 12px; border: none;">(2995.68)</td> <td style="padding-right: 12px; border: none;">(3031.21)</td> <td style="padding-right: 12px; border: none;">(2972.28)</td> <td style="padding-right: 12px; border: none;">(113.33)</td> <td style="padding-right: 12px; border: none;">(599.66)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Median Age</td> <td style="padding-right: 12px; border: none;">-38.56<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">-27.84<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">-28.06<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">-27.84<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-24.90<sup style="vertical-align: 0px;">*</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(10.89)</td> <td style="padding-right: 12px; border: none;">(10.94)</td> <td style="padding-right: 12px; border: none;">(11.28)</td> <td style="padding-right: 12px; border: none;">(11.02)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(9.40)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Percent Male</td> <td style="padding-right: 12px; border: none;">-115.67<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">-23.28</td> <td style="padding-right: 12px; border: none;">-23.21</td> <td style="padding-right: 12px; border: none;">-12.29</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(31.39)</td> <td style="padding-right: 12px; border: none;">(44.75)</td> <td style="padding-right: 12px; border: none;">(45.23)</td> <td style="padding-right: 12px; border: none;">(44.59)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;">Gini</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">43.01<sup style="vertical-align: 0px;">**</sup></td> <td style="padding-right: 12px; border: none;">43.71<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;">39.04<sup style="vertical-align: 0px;">*</sup></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">38.68<sup style="vertical-align: 0px;">**</sup></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(15.68)</td> <td style="padding-right: 12px; border: none;">(17.36)</td> <td style="padding-right: 12px; border: none;">(17.15)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(11.67)</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Poverty Rate</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">-0.88</td> <td style="padding-right: 12px; border: none;">-6.09</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(8.91)</td> <td style="padding-right: 12px; border: none;">(9.17)</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> </tr> <tr> <td style="padding-right: 12px; border: none;">Unemployment Rate</td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">25.47</td> <td style="padding-right: 12px; border: none;">45.89<sup style="vertical-align: 0px;">***</sup></td> <td style="padding-right: 12px; border: none;">23.02</td> </tr> <tr> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;"></td> <td style="padding-right: 12px; border: none;">(14.18)</td> <td style="padding-right: 12px; border: none;">(12.99)</td> <td style="padding-right: 12px; border: none;">(13.14)</td> </tr> <tr> <td style="border-top: 1px solid black;">R2</td> <td style="border-top: 1px solid black;">0.29</td> <td style="border-top: 1px solid black;">0.39</td> <td style="border-top: 1px solid black;">0.39</td> <td style="border-top: 1px solid black;">0.43</td> <td style="border-top: 1px solid black;">0.20</td> <td style="border-top: 1px solid black;">0.42</td> </tr> <tr> <td style="padding-right: 12px; border: none;">Adjusted R2</td> <td style="padding-right: 12px; border: none;">0.26</td> <td style="padding-right: 12px; border: none;">0.35</td> <td style="padding-right: 12px; border: none;">0.34</td> <td style="padding-right: 12px; border: none;">0.37</td> <td style="padding-right: 12px; border: none;">0.19</td> <td style="padding-right: 12px; border: none;">0.39</td> </tr> <tr> <td style="border-bottom: 2px solid black;">BIC (null)</td> <td style="border-bottom: 2px solid black;">-9.70</td> <td style="border-bottom: 2px solid black;">-13.34</td> <td style="border-bottom: 2px solid black;">-9.42</td> <td style="border-bottom: 2px solid black;">-9.02</td> <td style="border-bottom: 2px solid black;">-7.63</td> <td style="border-bottom: 2px solid black;">-16.28</td> </tr> <tr> <td style="padding-right: 12px; border: none;" colspan="8"><span style="font-size:0.8em"><sup style="vertical-align: 0px;">***</sup>p < 0.001; <sup style="vertical-align: 0px;">**</sup>p < 0.01; <sup style="vertical-align: 0px;">*</sup>p < 0.05</span></td> </tr> </table> ] ??? BIC prefers model 6, while adjusted `\(R^2\)` prefers model 7 and is indifferent between models 5 and 6. --- ## Model uncertainty -- - Given a set of potential independent variables, there is a very large number of potential models that could be chosen, even if we don't consider added complications like interaction or polynomial terms. -- - Generally, we select only one model from this full set. Even when we examine alternative models, we don't examine every possible set. -- - **Model averaging** techniques iteratively fit all or most models and use some algorithm to decide to average effects across these models. -- - The `BMA` package in R will do **Bayesian Model Averaging** which will provide an expected value for every coefficient across all possible models, as well as the probability a variable is in the model, and the top preferred models. --- ## Bayesian Model Averaging ```r library(BMA) model.bma <- bic.glm(crimes[,c("MedianAge","PctMale","PctLessHS","MedianIncomeHH", "Unemployment","Poverty", "Gini")], crimes$Violent, glm.family=gaussian) summary(model.bma) ``` ``` 29 models were selected Best 5 models (cumulative posterior probability = 0.5217 ): p!=0 EV SD model 1 model 2 model 3 model 4 model 5 Intercept 100 -54.3247 858.601 58.711 9.610 565.958 112.820 432.648 MedianAge 25.8 -3.2599 6.918 . . -13.550 . -11.076 PctMale 13.2 3.4006 14.069 . . . . . PctLessHS 49.2 8.0313 10.039 . 11.676 . 19.896 9.677 MedianIncomeHH 15.7 0.5853 2.228 . . . . . Unemployment 80.2 25.2981 16.451 36.220 24.508 36.452 . 26.702 Poverty 14.1 0.7423 7.776 . . . . . Gini 9.3 0.3648 4.671 . . . . . nVar 1 2 2 1 3 BIC -139.138 -138.340 -138.186 -137.665 -136.314 post prob 0.173 0.116 0.108 0.083 0.042 ```